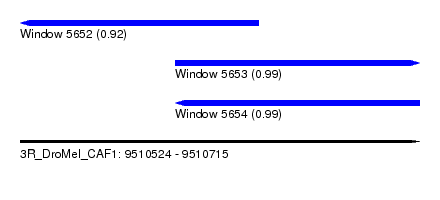
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,510,524 – 9,510,715 |
| Length | 191 |
| Max. P | 0.994588 |

| Location | 9,510,524 – 9,510,638 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 98.86 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9510524 114 - 27905053 CACAGACACAACGUCUGGCAAGUACAUCGCUUUUUGGGCCGUCUCUUUUGCGGUUCAUUCAAUAAAAUUAUUGCGAAUGGCAGGCGCAAAACAGAAUCGCAAAAAUUGCAGUGU ..(((((.....)))))......((((.((..(((..((..(((.(((((((.(((((((((((....))))..))))))..).))))))).)))...))..)))..)).)))) ( -30.70) >DroSec_CAF1 140218 114 - 1 CACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCCGUCUCUUUUGCGGUUCAUUCAAUAAAAUUAUUGCGAAUGGCAGGCGCAAAACAGAAUCGCAAAAAUUGCAGUGU .....((((.(((((((.((......((((....((((((((.......))))))))...((((....)))))))).)).)))))))...........(((.....))).)))) ( -32.40) >DroSim_CAF1 130354 114 - 1 CACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCCGUCUCUUUUGCGGUUCAUUCAAUAAAAUUAUUGCGAAUGGCAGGCGCAAAACAGAAUCGCAAAAAUUGCAGUGU .....((((.(((((((.((......((((....((((((((.......))))))))...((((....)))))))).)).)))))))...........(((.....))).)))) ( -32.40) >DroEre_CAF1 148073 114 - 1 CACAGACACAUCGUCUGGCAAGUACAUCGCUUUUUGGGCCGUCUCUUUUGCGGUUCAUUCAAUAAAAUUAUUGCGAAUGGUAGGCGCAAAACAGAAUCGCAAAAAUUGCAGUGU ..(((((.....)))))......((((.((..(((..((..(((.(((((((.(((((((((((....))))..))))))..).))))))).)))...))..)))..)).)))) ( -30.70) >DroYak_CAF1 141819 114 - 1 CACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCCGUCUCUUUUGCGGUUCAUUCAAUAAAAUUAUUGCGAAUGGUAGGCGCAAAACAGAAUCGCAAAAAUUGCAGUGU ..(((((.....)))))......((((.((..(((..((..(((.(((((((.(((((((((((....))))..))))))..).))))))).)))...))..)))..)).)))) ( -30.70) >consensus CACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCCGUCUCUUUUGCGGUUCAUUCAAUAAAAUUAUUGCGAAUGGCAGGCGCAAAACAGAAUCGCAAAAAUUGCAGUGU ..(((((.....)))))......((((.((..(((..((..(((.(((((((.(((((((((((....))))..))))))..).))))))).)))...))..)))..)).)))) (-30.70 = -30.70 + -0.00)


| Location | 9,510,598 – 9,510,715 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -32.32 |
| Energy contribution | -34.06 |
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9510598 117 + 27905053 GGCCCAAAAAGCGAUGUACUUGCCAGACGUUGUGUCUGUGUGCGUCCAAGUGUCUGUAGCGCGUUGUGUGUGUAUCGCGUAUGAAAAUACAUCAACUGUUGGGCAUCAACAAAA--AAA .((((((...((((((.....(((((((.....))))).))(((..(((((((.....)))).)))..))).))))))((((....))))........))))))..........--... ( -36.20) >DroSec_CAF1 140292 117 + 1 GGCCCAAAAAGCGAUGUACUUGCCAGACGCUGUGUCUGUGUGCGUCCGAGUGUCUGUAGCGCGUUGUGUGUGUAUCGCGUAUGAAAAUACAUCAACUGUUGGGCAUCAACAAAA--AAA .((((((...(((((((((....((((((((.((..((....))..))))))))))..((((...)))))))))))))((((....))))........))))))..........--... ( -38.40) >DroSim_CAF1 130428 119 + 1 GGCCCAAAAAGCGAUGUACUUGCCAGACGCUGUGUCUGUGUGCGUCCGAGUGUCUGUAGCGCGUUGUGUGUGUAUCGCGUAUGAAAAUACAUCAACUGUUGGGCAUCAACAAAAAAAAA .((((((...(((((((((....((((((((.((..((....))..))))))))))..((((...)))))))))))))((((....))))........))))))............... ( -38.40) >DroEre_CAF1 148147 105 + 1 GGCCCAAAAAGCGAUGUACUUGCCAGACGAUGUGUCUGUGUGCGUUCGACGGUCUGUAGC--GUUG--UGUGUAUCGCGUAUGAAAAUACA-----UGUUGGGCAUAAAAAAAA----- .((((((...(((((((((..(((((((.....))))).)).....(((((........)--))))--.)))))))))((((....)))).-----..))))))..........----- ( -35.70) >DroYak_CAF1 141893 112 + 1 GGCCCAAAAAGCGAUGUACUUGCCAGACGCUGUGUCUGUGUGCGUUCGACGGUCUGUAGCGCGUUG--UGUGUAUCGCGUAUGAAAAUACAUCAACUGUUGGGCAUAAAAAAAA----- .((((((...(((((((((..(((((((.....))))).))(((((((......)).)))))....--.)))))))))((((....))))........))))))..........----- ( -37.60) >consensus GGCCCAAAAAGCGAUGUACUUGCCAGACGCUGUGUCUGUGUGCGUCCGAGUGUCUGUAGCGCGUUGUGUGUGUAUCGCGUAUGAAAAUACAUCAACUGUUGGGCAUCAACAAAA__AAA .((((((...(((((((((....(((((.....)))))(((((...((......))..)))))......)))))))))((((....))))........))))))............... (-32.32 = -34.06 + 1.74)

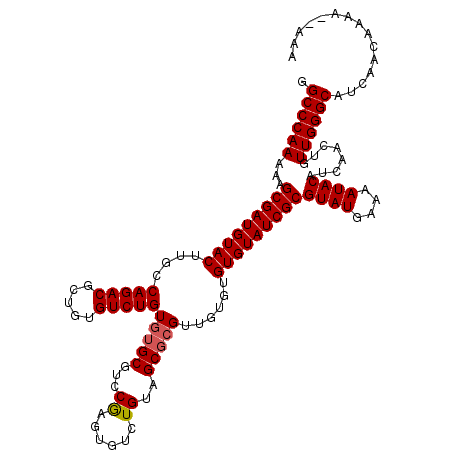
| Location | 9,510,598 – 9,510,715 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -26.46 |
| Energy contribution | -27.66 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9510598 117 - 27905053 UUU--UUUUGUUGAUGCCCAACAGUUGAUGUAUUUUCAUACGCGAUACACACACAACGCGCUACAGACACUUGGACGCACACAGACACAACGUCUGGCAAGUACAUCGCUUUUUGGGCC ...--..........((((((.((.((((((((((.(....................((((((........))).)))...(((((.....)))))).)))))))))).)).)))))). ( -32.00) >DroSec_CAF1 140292 117 - 1 UUU--UUUUGUUGAUGCCCAACAGUUGAUGUAUUUUCAUACGCGAUACACACACAACGCGCUACAGACACUCGGACGCACACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCC ...--..........((((((.((.((((((((((.(...((((............))))...........(((((((...........)))))))).)))))))))).)).)))))). ( -34.10) >DroSim_CAF1 130428 119 - 1 UUUUUUUUUGUUGAUGCCCAACAGUUGAUGUAUUUUCAUACGCGAUACACACACAACGCGCUACAGACACUCGGACGCACACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCC ...............((((((.((.((((((((((.(...((((............))))...........(((((((...........)))))))).)))))))))).)).)))))). ( -34.10) >DroEre_CAF1 148147 105 - 1 -----UUUUUUUUAUGCCCAACA-----UGUAUUUUCAUACGCGAUACACA--CAAC--GCUACAGACCGUCGAACGCACACAGACACAUCGUCUGGCAAGUACAUCGCUUUUUGGGCC -----..........((((((..-----.((((....))))(((((..((.--....--((....(.....)....))...(((((.....)))))....))..)))))...)))))). ( -24.90) >DroYak_CAF1 141893 112 - 1 -----UUUUUUUUAUGCCCAACAGUUGAUGUAUUUUCAUACGCGAUACACA--CAACGCGCUACAGACCGUCGAACGCACACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCC -----..........((((((.((.((((((((((.....((((.......--...)))).........((((.((((...........)))).)))))))))))))).)).)))))). ( -32.00) >consensus UUU__UUUUGUUGAUGCCCAACAGUUGAUGUAUUUUCAUACGCGAUACACACACAACGCGCUACAGACACUCGGACGCACACAGACACAGCGUCUGGCAAGUACAUCGCUUUUUGGGCC ...............((((((.((.((((((((((.(...((((............)))).....................(((((.....)))))).)))))))))).)).)))))). (-26.46 = -27.66 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:10 2006