
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,509,590 – 9,509,722 |
| Length | 132 |
| Max. P | 0.999337 |

| Location | 9,509,590 – 9,509,708 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -36.54 |
| Energy contribution | -36.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9509590 118 - 27905053 AUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUACAGCGAUAGUUGCAUAUGACGAGAAUGUGAGCUGUUAUCG ........(((((((((((....))))))))))).......((((((((.((((((.(((.(((((.......)))))((((....)))).......)))..)))).)).)))))))) ( -40.60) >DroSec_CAF1 139276 118 - 1 AUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUACAGCGAUAGUUGCAUUUGACGAGAAUGUGAGCUGUUAUCG ........(((((((((((....))))))))))).......((((((((...((.(((((.(((((.......)))))...))))).(..(((((.....)))))..))))))))))) ( -42.20) >DroSim_CAF1 129414 118 - 1 AUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUACAGCGAUAGUUGCAUUUGACGAGAAUGUGAGCUGUUAUCG ........(((((((((((....))))))))))).......((((((((...((.(((((.(((((.......)))))...))))).(..(((((.....)))))..))))))))))) ( -42.20) >DroEre_CAF1 147130 118 - 1 AUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGAGUAUCGAUACUUGGCAUUACAGUACAGCGAUAGUUGCAUAUGACGAAAAUAUGAGUUGUUAUCG ........(((((((((((....))))))))))).......((((((((.((((.(((((.((((.(......)))))...))))).)).(((((.......))))))).)))))))) ( -37.90) >DroYak_CAF1 140865 118 - 1 AUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGAGUAUCGAUACUUGGCAUUACAGUACAGCGAUAGUUGCAUAUGACGAGAAUAUGAGCUAUUAUCG ........(((((((((((....))))))))))).......((((((((.((((.(((((.((((.(......)))))...))))).)).(((((.......))))))).)))))))) ( -37.80) >consensus AUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUACAGCGAUAGUUGCAUAUGACGAGAAUGUGAGCUGUUAUCG ........(((((((((((....))))))))))).......((((((((.((...(((((.(((((.......)))))...)))))....(((((.......))))))).)))))))) (-36.54 = -36.94 + 0.40)


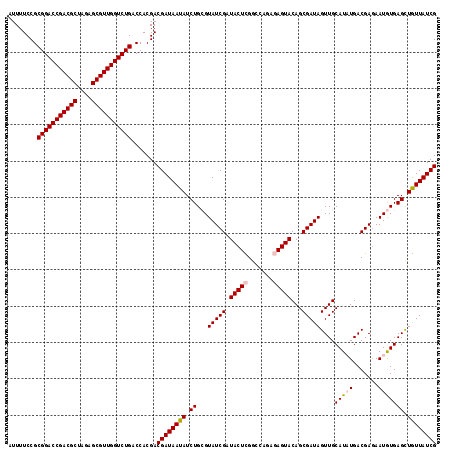
| Location | 9,509,630 – 9,509,722 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9509630 92 + 27905053 UACUCUCUGGCCGAGUAUCGAUACGCAGAUAUUAUCGUCGUGGUCAGACCAACGCUCUAGCGUCGGUCCGCGGAAAAUAGCGAAAAUUACAA .......((((((.....(((((.........)))))...))))))((((.((((....)))).))))(((........))).......... ( -25.70) >DroSec_CAF1 139316 92 + 1 UACUCUCUGGCCGAGUAUCGAUACGCAGAUAUUAUCGUCGUGGUCAGACCAACGCUCUAGCGUCGGUCCGCGGAAAAUAGCGAAAAUCACAA ........(((..((((((........))))))...)))(((((..((((.((((....)))).))))(((........)))...))))).. ( -27.40) >DroSim_CAF1 129454 92 + 1 UACUCUCUGGCCGAGUAUCGAUACGCAGAUAUUAUCGUCGUGGUCAGACCAACGCUCUAGCGUCGGUCCGCGGAAAAUAGCGAAAAUCACAA ........(((..((((((........))))))...)))(((((..((((.((((....)))).))))(((........)))...))))).. ( -27.40) >DroEre_CAF1 147170 92 + 1 UACUGUAAUGCCAAGUAUCGAUACUCAGAUAUUAUCGUCGUGGUCAGACCAACGCUCUAGCGUCGGUCCGCGGAAAAUAGCAAAAAUCACAA .((.(((((((..((((....))))..).)))))).)).(((((..((((.((((....)))).)))).((........))....))))).. ( -22.50) >DroYak_CAF1 140905 92 + 1 UACUGUAAUGCCAAGUAUCGAUACUCAGAUAUUAUCGUCGUGGUCAGACCAACGCUCUAGCGUCGGUCCGCGGAAAAUAGCAAAAAUCACAA .((.(((((((..((((....))))..).)))))).)).(((((..((((.((((....)))).)))).((........))....))))).. ( -22.50) >consensus UACUCUCUGGCCGAGUAUCGAUACGCAGAUAUUAUCGUCGUGGUCAGACCAACGCUCUAGCGUCGGUCCGCGGAAAAUAGCGAAAAUCACAA ........(((..((((((........))))))...)))(((((..((((.((((....)))).)))).((........))....))))).. (-23.02 = -23.26 + 0.24)



| Location | 9,509,630 – 9,509,722 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -25.74 |
| Energy contribution | -26.34 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9509630 92 - 27905053 UUGUAAUUUUCGCUAUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUA .....(((((((((........(((((((((((....))))))))))).......(((((.........)))))......)))..)))))). ( -27.60) >DroSec_CAF1 139316 92 - 1 UUGUGAUUUUCGCUAUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUA (((((.....(((........)))(((((((((....)))))))))....)))))(((((.........))))).(((((.......))))) ( -29.50) >DroSim_CAF1 129454 92 - 1 UUGUGAUUUUCGCUAUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUA (((((.....(((........)))(((((((((....)))))))))....)))))(((((.........))))).(((((.......))))) ( -29.50) >DroEre_CAF1 147170 92 - 1 UUGUGAUUUUUGCUAUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGAGUAUCGAUACUUGGCAUUACAGUA ((((((....(((((.......(((((((((((....))))))))))).......(((((.........))))).....))))))))))).. ( -29.20) >DroYak_CAF1 140905 92 - 1 UUGUGAUUUUUGCUAUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGAGUAUCGAUACUUGGCAUUACAGUA ((((((....(((((.......(((((((((((....))))))))))).......(((((.........))))).....))))))))))).. ( -29.20) >consensus UUGUGAUUUUCGCUAUUUUCCGCGGACCGACGCUAGAGCGUUGGUCUGACCACGACGAUAAUAUCUGCGUAUCGAUACUCGGCCAGAGAGUA (((((......((........))((((((((((....))))))))))...)))))(((((.........))))).(((((.......))))) (-25.74 = -26.34 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:05 2006