
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,502,560 – 9,502,701 |
| Length | 141 |
| Max. P | 0.996044 |

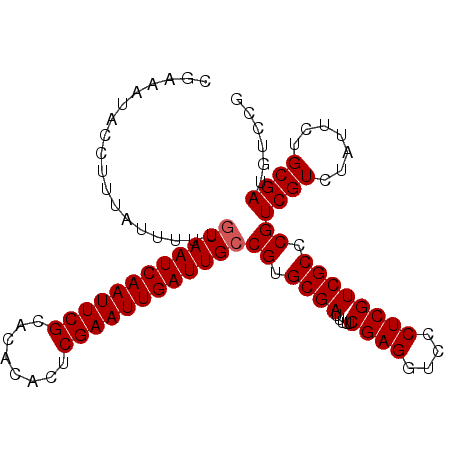
| Location | 9,502,560 – 9,502,662 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 97.84 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9502560 102 - 27905053 CGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACACACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGGUCCCUCGUCGCCCGUCGUCUAUUCUGCGAUGUCCG ..................((((((((((((........))))))))))))...((((.....((((....)))))))).((((((.......)))))).... ( -27.20) >DroSec_CAF1 132300 102 - 1 CGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACACACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGGUCCCUCGUCGCCCGUCGUCUAUUCUGCGAUGUCCG ..................((((((((((((........))))))))))))...((((.....((((....)))))))).((((((.......)))))).... ( -27.20) >DroSim_CAF1 122465 102 - 1 CGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACACACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGGUCCCUCGUCGCCCGUCGUCUAUUCUGCGAUGUCCG ..................((((((((((((........))))))))))))...((((.....((((....)))))))).((((((.......)))))).... ( -27.20) >DroEre_CAF1 140088 102 - 1 CGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACACACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGGUCCCUCGUCGCCCGUCGUUUAUUUUGCGAAGUCCG (((((((...........((((((((((((........))))))))))))((.((((.....((((....)))))))).)).....)))))))......... ( -27.00) >DroYak_CAF1 133850 102 - 1 CGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACACACUCGAAUUGAUUGGCGUGCGAUUUUUCGAGGUCCCUCGUCGCCCGUCGUUUAUUUUGCGAAGUCCG (((((((.............((((((((((........))))))))))((((.((((.....((((....)))))))).))))...)))))))......... ( -27.90) >consensus CGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACACACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGGUCCCUCGUCGCCCGUCGUCUAUUCUGCGAUGUCCG ..................((((((((((((........))))))))))))((.((((.....((((....)))))))).))((((.......))))...... (-25.22 = -25.42 + 0.20)


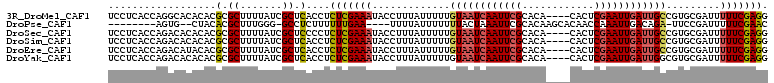
| Location | 9,502,595 – 9,502,701 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.85 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -14.86 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9502595 106 - 27905053 UCCUCACCAGGCACACACGCGCUUUUAUCGCUCACCUCUCGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACA----CACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGG .((((...((((.(....).))))..(((((.(.......((((((....)))))).((((((((((((....----....)))))))))))).).))))).....)))) ( -27.80) >DroPse_CAF1 135922 94 - 1 --------AGUG--CUACACGCUUUGGG-GCCUCUUUUUUGAA----UUUUAUUUUUUUACUAAAUUCGCACAAGCACAACCAAAUUGACAGA-UUCCGAUUUUUCGAAC --------.(((--((....(((....)-)).........(((----(((............)))))).....))))).......((((.(((-(....)))).)))).. ( -13.10) >DroSec_CAF1 132335 106 - 1 UCCUCACCAGACACACACGCGCUUUUAUCGCUCCCCUCUCGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACA----CACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGG .((((.............((((..................((((((....)))))).((((((((((((....----....)))))))))))).))))((....)))))) ( -24.80) >DroSim_CAF1 122500 106 - 1 UCCUCACCAGACACACACGCGCUUUUAUCGCUCACCUCUCGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACA----CACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGG .((((.............((((..................((((((....)))))).((((((((((((....----....)))))))))))).))))((....)))))) ( -24.80) >DroEre_CAF1 140123 106 - 1 UCCUCACCAGACAUACACGCGCUUUUAUCGCUCACCUCUCGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACA----CACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGG .((((.............((((..................((((((....)))))).((((((((((((....----....)))))))))))).))))((....)))))) ( -24.80) >DroYak_CAF1 133885 106 - 1 UCCUCACCAGACACACACGCGCUUUUAUCGCUCACCUCUCGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACA----CACUCGAAUUGAUUGGCGUGCGAUUUUUCGAGG .((((..........(((((((.......))...........................(((((((((((....----....)))))))))))))))).((....)))))) ( -24.30) >consensus UCCUCACCAGACACACACGCGCUUUUAUCGCUCACCUCUCGAAAUACCUUUAUUUUUGUAAUCAAUUCGCACA____CACUCGAAUUGAUUGCCGUGCGAUUUUUCGAGG ..................(.((.......)).)....(((((((.............((((((((((((............)))))))))))).........))))))). (-14.86 = -14.95 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:59 2006