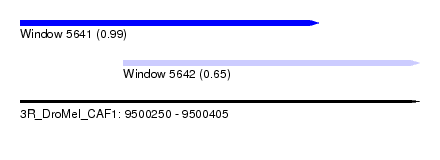
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,500,250 – 9,500,405 |
| Length | 155 |
| Max. P | 0.985229 |

| Location | 9,500,250 – 9,500,366 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -34.78 |
| Energy contribution | -34.82 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
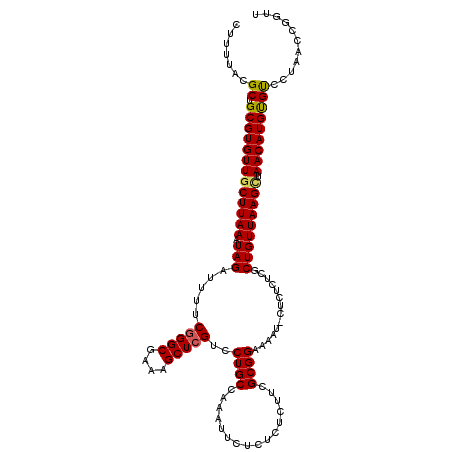
>3R_DroMel_CAF1 9500250 116 + 27905053 GGCAAUCGGAUGCUGCGGCGCGGGUGAAAAAAGUUUUAAACUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCGCUCUCUUCGCGGAAAA ((((...((((...((((((((((((..((((((.....)))))).)))).))))))))..............((((....))))))))))))...(((((.......)))))... ( -40.60) >DroSec_CAF1 130014 116 + 1 GGCAAUCGGAUGCUGCGGCGCGGGUGAUAAAAGUUUUAAACUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCUCUCUCUUCGCGGAAAA ((((...((((...((((((((((((.(((((((.....))))))))))).))))))))..............((((....))))))))))))....................... ( -38.80) >DroSim_CAF1 120187 116 + 1 GGCAAUCGGAUGCUGCGGCGCGGGUGAAAAAAGUUUUAAACUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCUCUCUCUUCGCGGAAAA ((((...((((...((((((((((((..((((((.....)))))).)))).))))))))..............((((....))))))))))))....................... ( -35.90) >DroEre_CAF1 137656 116 + 1 GGCAAUCGGAUGCUGCGGCGCGGGUGAAAAAAGUUUGAAACUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCUGGCGAAAGCUCGUCCUGCCAAAUUCGCUCUCUUCGCGGAAAA ((((...(((((..((((((((((((..((((((.....)))))).)))).))))))))...............(((....))))))))))))...(((((.......)))))... ( -38.30) >DroYak_CAF1 131437 115 + 1 GGCAAUCGGAUGCUGCGGCGCGGGUGAAAAAAGUUUAAAACUUUUGCGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAU-CUCUCUCCUCGCGGAAAA ((((...((((...((((((((((((..((((((.....)))))).)))).))))))))..............((((....))))))))))))....-.....(((....)))... ( -38.50) >consensus GGCAAUCGGAUGCUGCGGCGCGGGUGAAAAAAGUUUUAAACUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCUCUCUCUUCGCGGAAAA ((((...((((...((((((((((((..((((((.....)))))).)))).))))))))..............((((....))))))))))))..........(((....)))... (-34.78 = -34.82 + 0.04)


| Location | 9,500,290 – 9,500,405 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.73 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9500290 115 + 27905053 CUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCGCUCUCUUCGCGGAAAAU-CUCUCUCACUGUUAAGCUAACAUGUGUCCUAACCGGUU ..........(((((((((((((..((((((((((((....)))).............(((.......)))))))))-)).........)))))).)))))))............. ( -31.00) >DroSec_CAF1 130054 115 + 1 CUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCUCUCUCUUCGCGGAAAAU-CUCUCUCGCUGUUAAGCUAACAUGUGUCCUAACCGGUU ..........(((((((((((((..((((((((((((....)))))..((((................)))))))))-)).........)))))).)))))))............. ( -27.49) >DroSim_CAF1 120227 116 + 1 CUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCUCUCUCUUCGCGGAAAAUCCUCUCUCGCUGUUAAGCUAACAUGUGUCCUAACCGGUU ..........(((((((((((((.........(((((....)))))......................(((((........)).)))..)))))).)))))))............. ( -27.50) >DroEre_CAF1 137696 115 + 1 CUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCUGGCGAAAGCUCGUCCUGCCAAAUUCGCUCUCUUCGCGGAAAAU-CACUCUCGCUGUUAAGAUAACAUGCGUCGUAACCGGUU ...(((((.(((((((((((((....(((((((((.(((((((................)))...))))))))))))-)((.......)))))).))))))))).)))))...... ( -31.79) >DroYak_CAF1 131477 114 + 1 CUUUUGCGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAU-CUCUCUCCUCGCGGAAAAU-CUCUCUCGCUGUUAAGUUAACAUGCGCUCUAACCGGUU .......((.(((((((((((((..((((((.(((((....))))).......))))-))........(((((....-...)).)))..))))).))))))))))........... ( -30.40) >consensus CUUUUACGCUGCGUGUUGCUUAAAUAGAUUUUCGGGCGAAAGCUCGUCCUGCCAAAUUCUCUCUCUUCGCGGAAAAU_CUCUCUCGCUGUUAAGCUAACAUGUGUCCUAACCGGUU .......((.(((((((((((((.(((.....(((((....)))))..((((................))))..............))))))))).)))))))))........... (-25.85 = -25.73 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:57 2006