| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,497,762 – 9,497,858 |
| Length | 96 |
| Max. P | 0.978048 |

| Location | 9,497,762 – 9,497,858 |
|---|---|
| Length | 96 |
| Sequences | 6 |
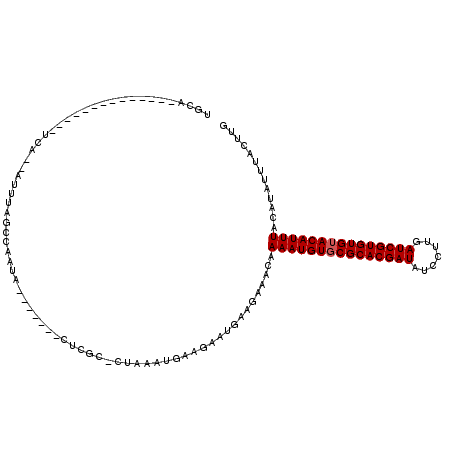
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.978048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9497762 96 - 27905053 AGGA--------------UCA--ACUCACCCAUUA-------CUCGC-CUAAAUGAAGAAUGAAGAAACAAAAUGUGCGCACGAUAUCCUUGAUCGUGUGUACAUUUACCUAUCUGCUUG (((.--------------...--.......((((.-------.(((.-.....)))..))))........(((((((((((((((.......))))))))))))))).)))......... ( -25.00) >DroVir_CAF1 168899 98 - 1 UGCA--------------UGU--UUUUAGCCAAUA-----ACCUCGC-CUAAAUGAAGCAUGAAGAAACAAAAUGUCCGCACGAUAUCCCUGAUCGUGUGUACAUUUACAUAUUUACUUG ..((--------------(((--((((((.(....-----.....).-)))...))))))))........((((((.((((((((.......)))))))).))))))............. ( -21.20) >DroPse_CAF1 130868 97 - 1 CACA----------------------UUGCCAAUAAAAUAACCUCGU-CUAAAUGAAGAAUGAAGAAACAAAAUGUGCGCACGAUAUCCUUGAUCGUGUGUACAUUUACGUAUCUGCUUG ....----------------------..((.............((((-((......)).))))(((.((.(((((((((((((((.......)))))))))))))))..)).)))))... ( -24.20) >DroGri_CAF1 168205 98 - 1 UGCA--------------UGU--UUCUAGCCAAUA-----ACCUCGC-CUAAAUGAAGCAUGAAGAAACAAAAUGUGCGCACGAUAUCCCUGAUCGUGUGUACAUUUACAUAUUUACUGG ..((--------------(((--(((.........-----.......-......))))))))........(((((((((((((((.......)))))))))))))))............. ( -27.65) >DroWil_CAF1 144857 99 - 1 AGAA--------------AUAGUAUUCAACAUAAC-------CUCGCCCUAAAUGAAGAAUGAAGAAACAAAAUGUGCGCACGAUAUCCUUGAUCGUGUGUACAUUUACAUAUUUACUUG ..((--------------(((.(((((..(((...-------..........)))..)))))........(((((((((((((((.......)))))))))))))))...)))))..... ( -23.12) >DroAna_CAF1 121744 109 - 1 UGCAUCCGCAUCCAUCAAUCA--AC-GACCACAUA-------CUCGC-CUAAAUGAAGAAUGAAGAAACAAAAUGUGCGCACGAUAUCCUUGAUCGUGUGUACAUUUACCUACUUGCUAG .(((((..(((.(.(((....--.(-((.......-------.))).-.....))).).)))..))....(((((((((((((((.......))))))))))))))).......)))... ( -25.80) >consensus UGCA______________UCA__AUUUAGCCAAUA_______CUCGC_CUAAAUGAAGAAUGAAGAAACAAAAUGUGCGCACGAUAUCCUUGAUCGUGUGUACAUUUACAUAUUUACUUG ......................................................................(((((((((((((((.......)))))))))))))))............. (-19.25 = -19.42 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:55 2006