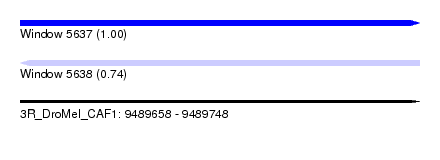
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,489,658 – 9,489,748 |
| Length | 90 |
| Max. P | 0.999886 |

| Location | 9,489,658 – 9,489,748 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -15.92 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9489658 90 + 27905053 UUAA-----------CUCUCUGCUAAAAACAUUCACAAUUUUAAUUACCAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUGA ....-----------.............(((((((((((..((((..........))))(((((.(((((....))))).)))))...))))))))))).. ( -16.00) >DroSec_CAF1 119701 90 + 1 UUAA-----------CUCUCUGCUAAAAACAUUCACAAUUUUAAUUACCAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUGA ....-----------.............(((((((((((..((((..........))))(((((.(((((....))))).)))))...))))))))))).. ( -16.00) >DroSim_CAF1 109805 90 + 1 UUAA-----------CUCUCUGCUAAAAACAUUCACAAUUUUAAUUACCAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUGA ....-----------.............(((((((((((..((((..........))))(((((.(((((....))))).)))))...))))))))))).. ( -16.00) >DroEre_CAF1 127292 89 + 1 UUAA-----------CUCUCUGCUAAAAACAUUCACAAUUUUAAUUACCAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUG- ....-----------.............(((((((((((..((((..........))))(((((.(((((....))))).)))))...))))))))))).- ( -16.00) >DroYak_CAF1 121763 89 + 1 UUAA-----------CUCUCUGCUAAAUACAUUCACAAUUUUAAUUACCAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUG- ....-----------............((((((((((((..((((..........))))(((((.(((((....))))).)))))...))))))))))))- ( -16.30) >DroAna_CAF1 114203 99 + 1 UUAAAAACAAAACAUCUCUCUGCUAA-AACAUUCACAACUUAAUUUAAAAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUG- ..........................-.((((((((((..((((...........))))(((((.(((((....))))).)))))....)))))))))).- ( -15.20) >consensus UUAA___________CUCUCUGCUAAAAACAUUCACAAUUUUAAUUACCAACAAUAUUAUAUAAAGAAUUGUCAAAUUCCUUAUAGCCAUUGUGAGUGUG_ ............................(((((((((((....................(((((.(((((....))))).)))))...))))))))))).. (-15.10 = -15.27 + 0.17)



| Location | 9,489,658 – 9,489,748 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.28 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9489658 90 - 27905053 UCACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUGGUAAUUAAAAUUGUGAAUGUUUUUAGCAGAGAG-----------UUAA (((((((.((((((..(((((((((((....)))))))))))...)))))).)))(((....)))))))......(((((.....)-----------)))) ( -17.00) >DroSec_CAF1 119701 90 - 1 UCACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUGGUAAUUAAAAUUGUGAAUGUUUUUAGCAGAGAG-----------UUAA (((((((.((((((..(((((((((((....)))))))))))...)))))).)))(((....)))))))......(((((.....)-----------)))) ( -17.00) >DroSim_CAF1 109805 90 - 1 UCACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUGGUAAUUAAAAUUGUGAAUGUUUUUAGCAGAGAG-----------UUAA (((((((.((((((..(((((((((((....)))))))))))...)))))).)))(((....)))))))......(((((.....)-----------)))) ( -17.00) >DroEre_CAF1 127292 89 - 1 -CACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUGGUAAUUAAAAUUGUGAAUGUUUUUAGCAGAGAG-----------UUAA -...((((((((((..(((((((((((....)))))))))))...))))))............((((.((.....)).)))).)))-----------)... ( -16.10) >DroYak_CAF1 121763 89 - 1 -CACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUGGUAAUUAAAAUUGUGAAUGUAUUUAGCAGAGAG-----------UUAA -.(((.(((((((...(((((((((((....)))))))))))...(((.....)))......))))))).)))..(((((.....)-----------)))) ( -16.30) >DroAna_CAF1 114203 99 - 1 -CACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUUUUAAAUUAAGUUGUGAAUGUU-UUAGCAGAGAGAUGUUUUGUUUUUAA -.(((.(((((((...(((((((((((....)))))))))))((((.(((....))))))).))))))).))).-..(((((((.....)))))))..... ( -17.40) >consensus _CACACUCACAAUGGCUAUAAGGAAUUUGACAAUUCUUUAUAUAAUAUUGUUGGUAAUUAAAAUUGUGAAUGUUUUUAGCAGAGAG___________UUAA .....(((((((((..(((((((((((....)))))))))))...))))))...................(((.....))))))................. (-15.28 = -15.28 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:53 2006