
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,480,952 – 9,481,128 |
| Length | 176 |
| Max. P | 0.898327 |

| Location | 9,480,952 – 9,481,054 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -27.69 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9480952 102 + 27905053 UGUUGGCUUGGGUGCCAUCGGAAUUACAUAGUGGGCCCAGCCGUUAGUGAAUGCCGAUGGCGGCAGGACGGAUUGGGCCAGAUAGGGAUUGUAGUUGGGAUU--- ..(..(((..(((.((((((......)......((((((((((((......(((((....))))).))))).))))))).))).)).)))..)))..)....--- ( -38.60) >DroVir_CAF1 152121 99 + 1 UGUUG---UUGGAAUUGUCGGUGCCACAUAUGAGGCCCAGCCAUUAAAGAAUGCCGAUGGUGGUAACGACGUUUGAGCCUGAUACGGAUUAUAGUUCGGAUU--- .((((---(((..((..((((.(((........))))).(.((((....)))).)))..))..)))))))((((((((.((((....))))..)))))))).--- ( -26.50) >DroSec_CAF1 110540 102 + 1 UGUUGGCUUGGAUGCCAUCGGAAUCACAUAGUGGGCCCAGCCGUUGGUGAAUGCCGAUGGCGGCAGAACGGACUGGGCCAGAUAGCGAUUGUAGUUGGGAUU--- ......((..(.(((.((((..(((.(.....)((((((((((((......(((((....))))).))))).))))))).)))..)))).))).)..))...--- ( -39.40) >DroSim_CAF1 100512 102 + 1 UGUUGGCUUGGAUGCCAUCGGAAUCACAUAGUGGGCCCAGCCGUUGGUGAAUGCCGAUGGCGGCAGAACGGACUGGGCCAGAUAGGGAUGGUAGUUGGGAUU--- ......((..(.(((((((.....(((...)))((((((((((((......(((((....))))).))))).))))))).......))))))).)..))...--- ( -43.60) >DroEre_CAF1 118093 102 + 1 UGUGGGCUUGGGUGCCACCGGAGUCACAUAGUGGGCCCAGCCGUGAGUGAAGGCUGAUGGCGGCAGGACGGACUGGGCCAGGUAGGGAUUGUAGUUGGCGUU--- .((.((((..(((.(((((.....(((...)))(((((((((((........((((....))))...)))).))))))).))).)).)))..)))).))...--- ( -39.30) >DroYak_CAF1 112421 105 + 1 UGUUGGCUUGGGUGCCACCGGAGUCACAUAGUGGGCCCAGCCAUGAGUGAAUGCCGAUGGCGGCAGGACGGACUGGGCCAGGUAGGGAUUGUAGUUGGCAUUGCU (((..(((..(((.(((((.....(((...)))(((((((((....((...(((((....)))))..)))).))))))).))).)).)))..)))..)))..... ( -42.20) >consensus UGUUGGCUUGGGUGCCAUCGGAAUCACAUAGUGGGCCCAGCCGUUAGUGAAUGCCGAUGGCGGCAGGACGGACUGGGCCAGAUAGGGAUUGUAGUUGGGAUU___ ..(..(((..(((.((((((......)......(((((((((((.......(((((....)))))..)))).))))))).))).)).)))..)))..)....... (-27.69 = -28.00 + 0.31)

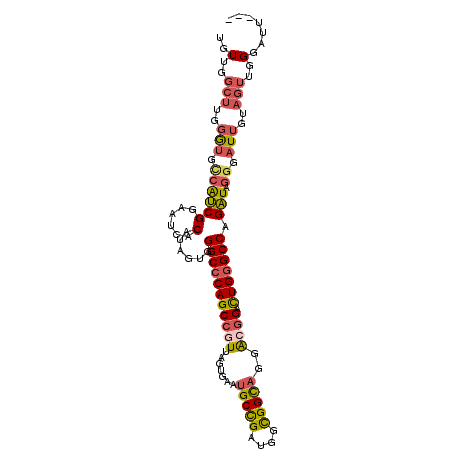

| Location | 9,480,952 – 9,481,054 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -20.93 |
| Energy contribution | -23.02 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9480952 102 - 27905053 ---AAUCCCAACUACAAUCCCUAUCUGGCCCAAUCCGUCCUGCCGCCAUCGGCAUUCACUAACGGCUGGGCCCACUAUGUAAUUCCGAUGGCACCCAAGCCAACA ---.......................((((((..((((..(((((....))))).......)))).))))))..............(.((((......)))).). ( -26.70) >DroVir_CAF1 152121 99 - 1 ---AAUCCGAACUAUAAUCCGUAUCAGGCUCAAACGUCGUUACCACCAUCGGCAUUCUUUAAUGGCUGGGCCUCAUAUGUGGCACCGACAAUUCCAA---CAACA ---..............(((((((.(((((((...(((((((((......)).......)))))))))))))).))))).))...............---..... ( -18.91) >DroSec_CAF1 110540 102 - 1 ---AAUCCCAACUACAAUCGCUAUCUGGCCCAGUCCGUUCUGCCGCCAUCGGCAUUCACCAACGGCUGGGCCCACUAUGUGAUUCCGAUGGCAUCCAAGCCAACA ---............((((((.....(((((((.(((((.(((((....)))))......))))))))))))......))))))..(.((((......)))).). ( -37.50) >DroSim_CAF1 100512 102 - 1 ---AAUCCCAACUACCAUCCCUAUCUGGCCCAGUCCGUUCUGCCGCCAUCGGCAUUCACCAACGGCUGGGCCCACUAUGUGAUUCCGAUGGCAUCCAAGCCAACA ---...........(((((.......(((((((.(((((.(((((....)))))......))))))))))))(((...))).....))))).............. ( -33.00) >DroEre_CAF1 118093 102 - 1 ---AACGCCAACUACAAUCCCUACCUGGCCCAGUCCGUCCUGCCGCCAUCAGCCUUCACUCACGGCUGGGCCCACUAUGUGACUCCGGUGGCACCCAAGCCCACA ---...(((.................(((((((.((((..((..((.....))...))...)))))))))))(((...))).....)))(((......))).... ( -28.00) >DroYak_CAF1 112421 105 - 1 AGCAAUGCCAACUACAAUCCCUACCUGGCCCAGUCCGUCCUGCCGCCAUCGGCAUUCACUCAUGGCUGGGCCCACUAUGUGACUCCGGUGGCACCCAAGCCAACA ......(((.................(((((((.((((..(((((....))))).......)))))))))))(((...))).....)))(((......))).... ( -31.90) >consensus ___AAUCCCAACUACAAUCCCUAUCUGGCCCAGUCCGUCCUGCCGCCAUCGGCAUUCACUAACGGCUGGGCCCACUAUGUGACUCCGAUGGCACCCAAGCCAACA ..........................(((((((.((((..(((((....))))).......)))))))))))..............(.((((......)))).). (-20.93 = -23.02 + 2.08)



| Location | 9,480,978 – 9,481,088 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -25.92 |
| Energy contribution | -27.09 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9480978 110 + 27905053 CAUAGUGGGCCCAGCCGUUAGUGAAUGCCGAUGGCGGCAGGACGGAUUGGGCCAGAUAGGGAUUGUAGUUGGGAUUGUUGUAAUUGUAGCUAUCAGCAGUGGCCGGCUGG .......((((((((((((......(((((....))))).))))).)))))))............(((((((.(((((((.............)))))))..))))))). ( -40.92) >DroVir_CAF1 152144 107 + 1 CAUAUGAGGCCCAGCCAUUAAAGAAUGCCGAUGGUGGUAACGACGUUUGAGCCUGAUACGGAUUAUAGUUCGGAUU---AUAGACUAAGUUAUCCAUAAUGGUCGGCUGG ..........(((((((((....)))((((...((((((((...(((((....((((.(((((....))))).)))---))))))...)))).))))..)))).)))))) ( -26.90) >DroSec_CAF1 110566 110 + 1 CAUAGUGGGCCCAGCCGUUGGUGAAUGCCGAUGGCGGCAGAACGGACUGGGCCAGAUAGCGAUUGUAGUUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGG ....((.((((((((((((......(((((....))))).))))).))))))).....)).....(((((((.(((((((((((....)))).)))))))..))))))). ( -45.80) >DroSim_CAF1 100538 110 + 1 CAUAGUGGGCCCAGCCGUUGGUGAAUGCCGAUGGCGGCAGAACGGACUGGGCCAGAUAGGGAUGGUAGUUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGG ..((((.((((..((((((((((..(((.(((.(((((((..(.((((..((((........)))))))).)..))))))).))))))..)))))))))))))).)))). ( -47.60) >DroEre_CAF1 118119 110 + 1 CAUAGUGGGCCCAGCCGUGAGUGAAGGCUGAUGGCGGCAGGACGGACUGGGCCAGGUAGGGAUUGUAGUUGGCGUUGUUGUAGUUGUAGCUAUCCAAAGUGGGCGGCUGG ..((((.(.(((((((((.(((....))).)))))((((.(((.(((.(.(((((.((.......)).))))).).)))...))).).)))........))))).)))). ( -37.20) >DroAna_CAF1 106158 110 + 1 CAUACGAGGCCCAGCCGUUAAUGAAGGCCGAUGGCGGGAGGACGGUCUGCGCCAGGUAGGGAUUGUAGGUGCUGCUGCUGUAGUUGUAGCUAUCCAGGGAGGCCGGUUGC ..(((((..(((.((((((......)))...((((((((......))).)))))))).))).)))))((((((((.((....)).)))))..((....)).)))...... ( -33.90) >consensus CAUAGUGGGCCCAGCCGUUAGUGAAUGCCGAUGGCGGCAGGACGGACUGGGCCAGAUAGGGAUUGUAGUUGGGAUUGUUGUAGUUGUAGCUAUCAACAGUGGCCGGCUGG .......((((((((((((......(((((....))))).))))).)))))))............(((((((.......(((((....))))).........))))))). (-25.92 = -27.09 + 1.17)



| Location | 9,480,978 – 9,481,088 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -15.71 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9480978 110 - 27905053 CCAGCCGGCCACUGCUGAUAGCUACAAUUACAACAAUCCCAACUACAAUCCCUAUCUGGCCCAAUCCGUCCUGCCGCCAUCGGCAUUCACUAACGGCUGGGCCCACUAUG (((((((((((.....(((((..............................))))))))))..........(((((....))))).........)))))).......... ( -27.81) >DroVir_CAF1 152144 107 - 1 CCAGCCGACCAUUAUGGAUAACUUAGUCUAU---AAUCCGAACUAUAAUCCGUAUCAGGCUCAAACGUCGUUACCACCAUCGGCAUUCUUUAAUGGCUGGGCCUCAUAUG ((((((((((..(((((((....(((((...---.....).))))..)))))))...)).))....((((..........))))..........)))))).......... ( -27.00) >DroSec_CAF1 110566 110 - 1 CCAGCCGGCCACCGCUGAUAGCUACAACUACAACAAUCCCAACUACAAUCGCUAUCUGGCCCAGUCCGUUCUGCCGCCAUCGGCAUUCACCAACGGCUGGGCCCACUAUG ..(((.(....).)))((((((............................)))))).(((((((.(((((.(((((....)))))......))))))))))))....... ( -34.39) >DroSim_CAF1 100538 110 - 1 CCAGCCGGCCACCGCUGAUAGCUACAACUACAACAAUCCCAACUACCAUCCCUAUCUGGCCCAGUCCGUUCUGCCGCCAUCGGCAUUCACCAACGGCUGGGCCCACUAUG .((((.(....).))))........................................(((((((.(((((.(((((....)))))......))))))))))))....... ( -32.10) >DroEre_CAF1 118119 110 - 1 CCAGCCGCCCACUUUGGAUAGCUACAACUACAACAACGCCAACUACAAUCCCUACCUGGCCCAGUCCGUCCUGCCGCCAUCAGCCUUCACUCACGGCUGGGCCCACUAUG ..(((..((......))...)))..................................(((((((.((((..((..((.....))...))...)))))))))))....... ( -24.20) >DroAna_CAF1 106158 110 - 1 GCAACCGGCCUCCCUGGAUAGCUACAACUACAGCAGCAGCACCUACAAUCCCUACCUGGCGCAGACCGUCCUCCCGCCAUCGGCCUUCAUUAACGGCUGGGCCUCGUAUG ((..((((.....))))...(((........))).))...............(((..(((.(((.((((......(((...)))........))))))).)))..))).. ( -27.64) >consensus CCAGCCGGCCACCGCGGAUAGCUACAACUACAACAAUCCCAACUACAAUCCCUAUCUGGCCCAGUCCGUCCUGCCGCCAUCGGCAUUCACUAACGGCUGGGCCCACUAUG .........................................................(((((((.((((..(((((....))))).......)))))))))))....... (-15.71 = -17.60 + 1.89)



| Location | 9,481,018 – 9,481,128 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -29.12 |
| Energy contribution | -28.02 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9481018 110 + 27905053 GACGGAUUGGGCCAGAUAGGGAUUGUAGUUGGGAUUGUUGUAAUUGUAGCUAUCAGCAGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGAACCGAGCUAAAUAUGGGGA ..(((.(((((((............(((((((.(((((((.............)))))))..))))))).(((........)))))))))).)))............... ( -33.92) >DroPse_CAF1 113828 107 + 1 GCUUGACUGUGCCAAAAACGGAUUGAAGUGUGGAUUGUAGCAGUU---UCUGUCCAUCACGGCUGGCUGCGGCGGCACGUAGCCGGCGCGGGCCGCACCGCCCAUGGGAA ((..((((((.(((...((........)).)))......))))))---...))((((....((((((((((......))))))))))..((((......))))))))... ( -40.80) >DroSec_CAF1 110606 110 + 1 AACGGACUGGGCCAGAUAGCGAUUGUAGUUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGA ..(((.(((((((............(((((((.(((((((((((....)))).)))))))..))))))).(((........)))))))))).)))............... ( -38.80) >DroSim_CAF1 100578 110 + 1 AACGGACUGGGCCAGAUAGGGAUGGUAGUUGGGAUUGUUGUAGUUGUAGCUAUCAGCGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGU ......((.(.((((.......(((((((((.(((((...))))).))))))))).(((((((((((((.(......).)))))))))...))))..))))...).)).. ( -40.80) >DroEre_CAF1 118159 110 + 1 GACGGACUGGGCCAGGUAGGGAUUGUAGUUGGCGUUGUUGUAGUUGUAGCUAUCCAAAGUGGGCGGCUGGGGCGGAAUGUAGCCGGCCCGGAUCGAGCUGGAAACGGGGA ..(((.(((((((.......(((....)))(((...(((...(((.(((((..((.....))..))))).)))..)))...)))))))))).)))..(((....)))... ( -41.00) >DroAna_CAF1 106198 110 + 1 GACGGUCUGCGCCAGGUAGGGAUUGUAGGUGCUGCUGCUGUAGUUGUAGCUAUCCAGGGAGGCCGGUUGCGGCGGAAUGUAGCCGGCGCGGGCGGCUCCGCCCACGGGGA ..(.(((((((((.(((..((((.((....(((((....)))))....)).))))......)))(((((((......)))))))))))))))).)(((((....))))). ( -49.20) >consensus GACGGACUGGGCCAGAUAGGGAUUGUAGUUGGGAUUGUUGUAGUUGUAGCUAUCAACGGUGGCCGGCUGGGGCGGAAUGUAGCCGGCUCGGACCGAGCUGGAAAUGGGGA ..(((.(((((((...........(((((((.(((((...))))).)))))))...........(((((.(......).)))))))))))).)))..(((....)))... (-29.12 = -28.02 + -1.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:50 2006