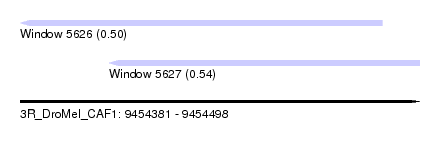
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,454,381 – 9,454,498 |
| Length | 117 |
| Max. P | 0.536251 |

| Location | 9,454,381 – 9,454,487 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9454381 106 - 27905053 GAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCACUCACUCAC--ACAGACAGCAACCCUCAC ........(((((.(((((((((((.........................)))))))))))))))).(((((....))......((..--...))..)))........ ( -27.71) >DroPse_CAF1 85944 94 - 1 GCAAAAUAAAACCCACGUCUCGGCAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGCGGCGCGUUUUAUGCGCG----CGCUCGCUCGCACACA--CA--------CAC .....(((((((...((((.(((((.........................))))).)))).)))))))(((.(----(....)).))).....--..--------... ( -20.81) >DroSec_CAF1 83973 108 - 1 GAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCACUCACUCACACACAGACGGCAACGCUCAC ........(((((.(((((((((((.........................)))))))))))))))).((((....))))..............((((....)).)).. ( -30.51) >DroSim_CAF1 73257 108 - 1 GAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCACUCACUCACACACAGACAGCAACGCUCAC ........(((((.(((((((((((.........................))))))))))))))))..(((...((((..((...........))..))))))).... ( -29.01) >DroWil_CAF1 92520 104 - 1 GAGAAAUAAAAACUACAACGCGGCAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGUGGCGCGUUUUAUGCGCACUUACACAAACACACACACAUACA----CCCGUAU .............(((...((((((............)))))).(((.......)))((((((...)))))).........................----...))). ( -17.50) >DroYak_CAF1 86052 106 - 1 GAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCACUCACUCAC--ACAGGCACCAAUCCUCAC ........(((((.(((((((((((.........................)))))))))))))))).(((((....)).....((...--..)))))........... ( -27.21) >consensus GAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCACUCACUCACACACAGACAGCAACCCUCAC ................(((((((((.........................)))))))))((((.....)))).................................... (-18.46 = -18.74 + 0.28)

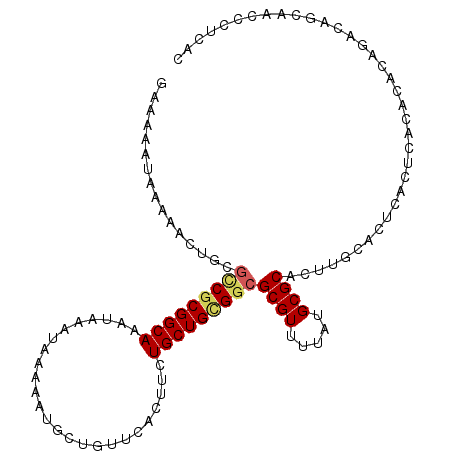

| Location | 9,454,407 – 9,454,498 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.86 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9454407 91 - 27905053 GAAUGGGCUAAGAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCAC ......((.(((....((((((...(((((((((((.........................))))))))))).)))))).....))))).. ( -27.01) >DroVir_CAF1 126536 87 - 1 ---UAAAU-AAAAAAAGGCAAAACUGCGCCGCAGAAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUCAAC ---.....-........(((((((.(((((((((.............................)))))))))))))..))).......... ( -22.15) >DroEre_CAF1 91769 91 - 1 GAAUGUGCUAAGAAAAAUAAAAACUGCGCCACGGCAAAUAAAUAAAAAUGCUGUUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCAC ....((((.(((....((((((...(((((.(((((.........................))))).))))).)))))).....))))))) ( -24.21) >DroWil_CAF1 92544 86 - 1 -----AGCAAAGAGAAAUAAAAACUACAACGCGGCAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGUGGCGCGUUUUAUGCGCACUUACAC -----((((((((....)............((((((............))))))...))).))))...((((((...))))))........ ( -18.00) >DroAna_CAF1 78951 91 - 1 GCAUGGAAAAAGAAAAAUAAAAACUGCACCGCUGCAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGCGCCGCGUUUUAUGCGCACUUGCAC ((((....................((((....))))........(((((((.((.(((....).)).)).)))))))))))((....)).. ( -17.60) >DroPer_CAF1 87677 87 - 1 GCACCAGCAAAGCAAAAUAAAACCCACGUCUCGGCAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGCGGCGCGUUUUAUGCGCG----CGC ((.(((((((((.........((....))..(((((............)))))....))).))))).)((((((.....)))))----))) ( -19.70) >consensus G_AUGAGCAAAGAAAAAUAAAAACUGCGCCGCGGCAAAUAAAUAAAAAUGCUGCUCACUUCUGCUGCGGCGCGUUUUAUGCGCACUUGCAC ...........................(((((((((.........................)))))))))((((.....))))........ (-13.80 = -14.86 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:43 2006