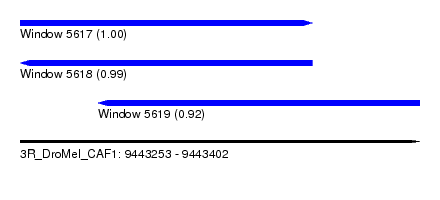
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,443,253 – 9,443,402 |
| Length | 149 |
| Max. P | 0.999915 |

| Location | 9,443,253 – 9,443,362 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.30 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9443253 109 + 27905053 AAUAAAGUCAUCACUAUUUGCAUGCACCGCAUGUUGGGAAUCCUUAAUAAUUUACAAUGGCAUGCCGUCAACUAAUUGUGGUGUUUGGGCUUCUUAGUUGACUUUGUUA ((((((((((..((((...((..(((((((((((((((....))))))).......((((....))))........))))))))....))....)))))))))))))). ( -33.40) >DroSec_CAF1 72833 109 + 1 AAUAAAGUCAUCACUAUUUGCAUGCACCGCAUGUUGGGAAUCCUUAAUAAUUCACAAUGGCAUGCCGUCAACUAAUUGUGGUGUUUGGGAUUCUUAGUUGACUUUGUAA .(((((((((.(.......((((((...))))))((((((((((.((((..((((((((((.....)))).....)))))))))).))))))))))).))))))))).. ( -35.00) >DroSim_CAF1 61692 109 + 1 AAUAAAGUCAUCACUAUUUGCAUGCGCCGCAUGUUGGGAAUCCUUAAUAAUUCACAAUGGCAUGCCGUCAACUAAUUGUGGUGUUUGGGCUUCUUAGUUGACUUUGUAA .(((((((((..((((...((..((((((((.(((((((((........))))...((((....))))...)))))))))))))....))....))))))))))))).. ( -33.40) >DroEre_CAF1 81026 109 + 1 AAUAAAGUCAUCACUAUUUGCAUGCACCGCAUGUGGGGAAUCCUUAAUAAUUCACAAUGGCAUGCCGUCAACUAAUUGCGGUGUUUGGGCUUCUUAGUUGACUUUGUAA .(((((((((..((((...((..((((((((((((((..((.....))..))))))((((....))))........))))))))....))....))))))))))))).. ( -34.50) >DroYak_CAF1 75233 109 + 1 AAUAAAGUCAUCACUAUUUGCAUGCACCGCAUGUUGGGAAUCCUUAAUAAUUCACAAUGGCAUGCCGUCAACUAAUUGUGGUGUUUGGGCUUCUUAGUUGACUUUGUAA .(((((((((..((((...((..((((((((.(((((((((........))))...((((....))))...)))))))))))))....))....))))))))))))).. ( -33.80) >consensus AAUAAAGUCAUCACUAUUUGCAUGCACCGCAUGUUGGGAAUCCUUAAUAAUUCACAAUGGCAUGCCGUCAACUAAUUGUGGUGUUUGGGCUUCUUAGUUGACUUUGUAA .(((((((((..((((...((..((((((((.(((((((((........))))...((((....))))...)))))))))))))....))....))))))))))))).. (-32.38 = -32.30 + -0.08)


| Location | 9,443,253 – 9,443,362 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9443253 109 - 27905053 UAACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUAAAUUAUUAAGGAUUCCCAACAUGCGGUGCAUGCAAAUAGUGAUGACUUUAUU .......(((((((((..................))))))))).((((((.((((((.........((....))....))))))))))))................... ( -22.09) >DroSec_CAF1 72833 109 - 1 UUACAAAGUCAACUAAGAAUCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUGCGGUGCAUGCAAAUAGUGAUGACUUUAUU ((((...(((((((((..................))))))))).((((((.((((((((((......))))).......))))))))))).....)))).......... ( -25.58) >DroSim_CAF1 61692 109 - 1 UUACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUGCGGCGCAUGCAAAUAGUGAUGACUUUAUU ((((...(((((((((..................))))))))).(((((((..((((((((......)))))...)))....).)))))).....)))).......... ( -24.77) >DroEre_CAF1 81026 109 - 1 UUACAAAGUCAACUAAGAAGCCCAAACACCGCAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCCACAUGCGGUGCAUGCAAAUAGUGAUGACUUUAUU ((((...(((((((((...((.........))..))))))))).((((((.((((((((((......))))).......))))))))))).....)))).......... ( -27.61) >DroYak_CAF1 75233 109 - 1 UUACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUGCGGUGCAUGCAAAUAGUGAUGACUUUAUU ((((...(((((((((..................))))))))).((((((.((((((((((......))))).......))))))))))).....)))).......... ( -25.58) >consensus UUACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUGCGGUGCAUGCAAAUAGUGAUGACUUUAUU ((((...(((((((((..................))))))))).(((((((..((((((((......)))))...)))....).)))))).....)))).......... (-23.33 = -23.73 + 0.40)


| Location | 9,443,282 – 9,443,402 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -19.99 |
| Energy contribution | -19.83 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9443282 120 - 27905053 UAGGAGCAAAUGAUUAUUAAUUUAUGCUCAGUUACUGCAAUAACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUAAAUUAUUAAGGAUUCCCAACAUG ...(((((...(((.....)))..))))).(((..((((((......(((((((((..................)))))))))((....)))))))).........((....)))))... ( -23.77) >DroSec_CAF1 72862 120 - 1 UAGGACCAAAUGAUUAUUAAUUUAUGCUCAAUUACUGCAAUUACAAAGUCAACUAAGAAUCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUG ..(((((.(((((((.........(((.........)))..(((((.(((((((((..................)))))))))((....)).))))))))))))..))..)))....... ( -22.27) >DroSim_CAF1 61721 120 - 1 UAGGAGCAAAUGAUUAUUAAUUUAUGCUCAAUUACUGCAAUUACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUG ...(((((...(((.....)))..)))))...........((((((.(((((((((..................)))))))))((....)).))))))........((....))...... ( -23.07) >DroEre_CAF1 81055 116 - 1 ----UGCUAAUGAUUAUUAAUUUAUGCUCAAUUACUGCAAUUACAAAGUCAACUAAGAAGCCCAAACACCGCAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCCACAUG ----...((((((((.........(((.........)))..(((((.(((((((((...((.........))..)))))))))((....)).))))))))))))).((.....))..... ( -23.60) >DroYak_CAF1 75262 120 - 1 UAGGAGCAAAUGAUUAUUAAUUUAUGCUCAAUUACUGCAAUUACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUG ...(((((...(((.....)))..)))))...........((((((.(((((((((..................)))))))))((....)).))))))........((....))...... ( -23.07) >consensus UAGGAGCAAAUGAUUAUUAAUUUAUGCUCAAUUACUGCAAUUACAAAGUCAACUAAGAAGCCCAAACACCACAAUUAGUUGACGGCAUGCCAUUGUGAAUUAUUAAGGAUUCCCAACAUG .........(((.(((.((((((.(((.........)))...((((.(((((((((..................)))))))))((....)).)))))))))).)))((....))..))). (-19.99 = -19.83 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:35 2006