
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,429,461 – 9,429,554 |
| Length | 93 |
| Max. P | 0.850759 |

| Location | 9,429,461 – 9,429,554 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -19.62 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
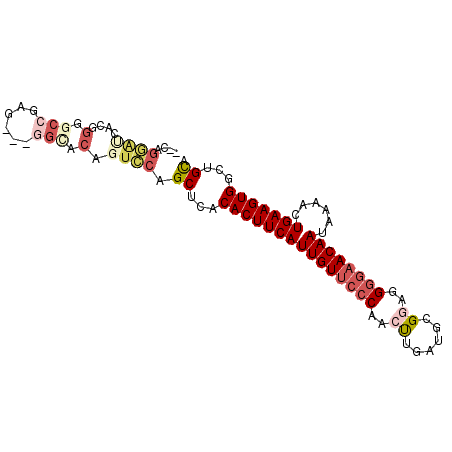
>3R_DroMel_CAF1 9429461 93 + 27905053 --CAGGAUCACGGGACCGAGUCCAGCACAGUCCAGCUCACACUUCAUUGUUCCCAACUUGAUGCGAAGGGGAACAAUAAAACUGAAGUGGCUGCA --..((((....((((...))))......)))).((...(((((((((((((((..(((......)))))))))))......)))))))...)). ( -30.20) >DroSec_CAF1 59091 90 + 1 --CAGGAUCACGGUGCCGAG---GGCACAGUCCAGCUCACACUUCAUUGUUCGCAACUUGAUGCGGAGGGGAACAAUAUAACUGAAGUGGCUGCA --..((((....(((((...---))))).)))).((...(((((((...((((((......)))))).(....)........)))))))...)). ( -30.00) >DroEre_CAF1 65228 92 + 1 GGCAGGAUCAUGGGGCCGAG---GGCACAGUCCAGCUCACACUUCAUUGUUCCCAACUUGAUGCGGAGGGGAACAAUAAAACUGAAGUGGCUGCC (((((.......((((...(---(((...)))).)))).(((((((((((((((..((......))..))))))))......))))))).))))) ( -35.50) >DroYak_CAF1 61319 90 + 1 --CAGGACCACGGGGCCGAG---GGCGCAGUCCAGCUCACACUUCAUUGUUCCCAACUUGAUGCGGAGGGGAACAAUAAAACUGAAGUGGCUGCA --..((((....(.(((...---))).).)))).((...(((((((((((((((..((......))..))))))))......)))))))...)). ( -33.70) >DroAna_CAF1 55865 75 + 1 -----------------GAG---GGCACUGGCGAGCUCACACUUCAUUGUUCCCAGCUGGACGAGAAGGGAAACAAUAAAACUGAAGUGGCUGCA -----------------..(---(((........)))).(((((((((((((((..((.....))..))).)))))......)))))))...... ( -21.10) >DroPer_CAF1 58614 86 + 1 ----GAGCCACGGAGCCAGG---GGU-CCUUUCGGCUCACACUUCAUUGUUCCCGGCCAUA-GCGGAGGGGCACAAUAAAACUGAAGUGGCUGUG ----.((((((.((((((((---...-.)))..))))).......(((((.(((..((...-..))..))).))))).........))))))... ( -32.30) >consensus __CAGGAUCACGGGGCCGAG___GGCACAGUCCAGCUCACACUUCAUUGUUCCCAACUUGAUGCGGAGGGGAACAAUAAAACUGAAGUGGCUGCA ....((((....(.(((......))).).)))).((...(((((((((((((((..((......))..))))))))......)))))))...)). (-19.62 = -21.52 + 1.89)

| Location | 9,429,461 – 9,429,554 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9429461 93 - 27905053 UGCAGCCACUUCAGUUUUAUUGUUCCCCUUCGCAUCAAGUUGGGAACAAUGAAGUGUGAGCUGGACUGUGCUGGACUCGGUCCCGUGAUCCUG-- .((((((((((((.((((((((((((((((......)))..))))))))))))))).))).))).))))...((((.((....)).).)))..-- ( -31.50) >DroSec_CAF1 59091 90 - 1 UGCAGCCACUUCAGUUAUAUUGUUCCCCUCCGCAUCAAGUUGCGAACAAUGAAGUGUGAGCUGGACUGUGCC---CUCGGCACCGUGAUCCUG-- ..(((((((((((......((((((......(((......))))))))))))))))...)))).((.(((((---...))))).)).......-- ( -25.30) >DroEre_CAF1 65228 92 - 1 GGCAGCCACUUCAGUUUUAUUGUUCCCCUCCGCAUCAAGUUGGGAACAAUGAAGUGUGAGCUGGACUGUGCC---CUCGGCCCCAUGAUCCUGCC (((((((((((((.(((((((((((((..(........)..))))))))))))))).))).))).....(((---...))).........))))) ( -29.90) >DroYak_CAF1 61319 90 - 1 UGCAGCCACUUCAGUUUUAUUGUUCCCCUCCGCAUCAAGUUGGGAACAAUGAAGUGUGAGCUGGACUGCGCC---CUCGGCCCCGUGGUCCUG-- .((((((((((((.(((((((((((((..(........)..))))))))))))))).))).))).))))..(---(.((....)).)).....-- ( -32.90) >DroAna_CAF1 55865 75 - 1 UGCAGCCACUUCAGUUUUAUUGUUUCCCUUCUCGUCCAGCUGGGAACAAUGAAGUGUGAGCUCGCCAGUGCC---CUC----------------- .((((((((.....((((((((((.(((..((.....))..))))))))))))).))).))).)).......---...----------------- ( -20.30) >DroPer_CAF1 58614 86 - 1 CACAGCCACUUCAGUUUUAUUGUGCCCCUCCGC-UAUGGCCGGGAACAAUGAAGUGUGAGCCGAAAGG-ACC---CCUGGCUCCGUGGCUC---- ...((((((.....(((((((((.(((..((..-...))..))).))))))))).(.(((((...(((-...---))))))))))))))).---- ( -31.60) >consensus UGCAGCCACUUCAGUUUUAUUGUUCCCCUCCGCAUCAAGUUGGGAACAAUGAAGUGUGAGCUGGACUGUGCC___CUCGGCCCCGUGAUCCUG__ ..(((((((.....(((((((((((((..(........)..))))))))))))).))).))))................................ (-19.21 = -19.68 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:27 2006