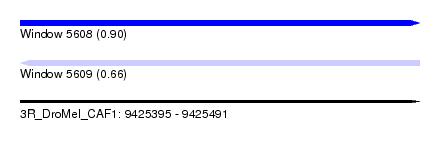
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,425,395 – 9,425,491 |
| Length | 96 |
| Max. P | 0.900170 |

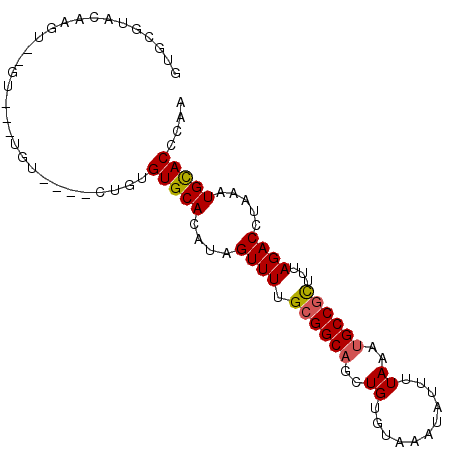
| Location | 9,425,395 – 9,425,491 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -20.01 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.49 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9425395 96 + 27905053 UGGGGUGCAUUUAGGUCUAAAGCGGCAUUUAAAAUAUUUACACAGCUGCCGCAAAACUAUGUGCACACAGACAAACAAAAGCACACUUGUACGCAC ..(.(((((............((((((...................)))))).......(((((................)))))..))))).).. ( -21.10) >DroPse_CAF1 54244 78 + 1 UAGGGUGCAUUUAGGUCUAAAGCGGCAUUUAAAAUAUUUACACAGCUGCCGCAAAACUAUGUGCACACAA----AUA--------------CACAC ....((((((.(((.......((((((...................))))))....))).))))))....----...--------------..... ( -19.31) >DroEre_CAF1 61009 90 + 1 UUGGGUGCAUUUAGGUCUAAAGCGGCAUUUAAAAUAUUUACACAGCUGCCGCAAAACUAUGUGCACACGC----ACAC--ACACACUUGUACGCAC .((.(((((.....((.....((((((...................)))))).......(((((....))----))).--)).....))))).)). ( -23.51) >DroWil_CAF1 59111 88 + 1 UUGAGUACAUUUAAGUCUAAAAGGGCGUUUAAAAUAUUUACACAGAUGCCGCAAAACUAUGUGCACCCAG----ACAC--AC--ACACACACCCAC ..............((((.....(((((((.............)))))))(((........)))....))----))..--..--............ ( -13.32) >DroYak_CAF1 57086 88 + 1 UGGUGUGCAUUUGGGUCUAAAGCGGCAUUUAAAAUAUUUACACAGCUGCCGCAAAACUAUGUGCACACUC----AC----ACACACUUGUACGCAC .(((((((((.(((.......((((((...................))))))....))).))))))))).----..----................ ( -23.51) >DroPer_CAF1 54894 78 + 1 UAGGGUGCAUUUAGGUCUAAAGCGGCAUUUAAAAUAUUUACACAGCUGCCGCAAAACUAUGUGCACACAA----AUA--------------UACAC ....((((((.(((.......((((((...................))))))....))).))))))....----...--------------..... ( -19.31) >consensus UAGGGUGCAUUUAGGUCUAAAGCGGCAUUUAAAAUAUUUACACAGCUGCCGCAAAACUAUGUGCACACAA____ACA___AC__ACUUGUACGCAC ....((((((.(((.......((((((...................))))))....))).)))))).............................. (-16.10 = -16.49 + 0.39)

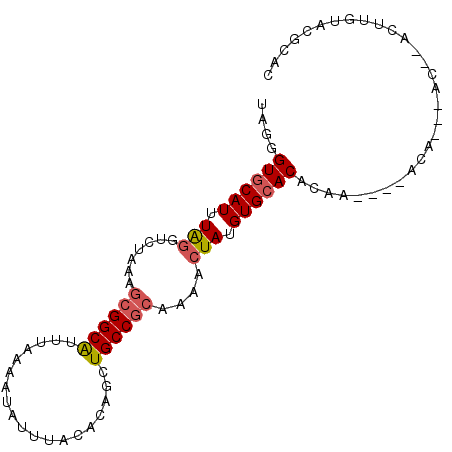
| Location | 9,425,395 – 9,425,491 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -15.86 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9425395 96 - 27905053 GUGCGUACAAGUGUGCUUUUGUUUGUCUGUGUGCACAUAGUUUUGCGGCAGCUGUGUAAAUAUUUUAAAUGCCGCUUUAGACCUAAAUGCACCCCA ((((((....((((((................)))))).((((.((((((..((...........))..))))))...))))....)))))).... ( -25.69) >DroPse_CAF1 54244 78 - 1 GUGUG--------------UAU----UUGUGUGCACAUAGUUUUGCGGCAGCUGUGUAAAUAUUUUAAAUGCCGCUUUAGACCUAAAUGCACCCUA (((((--------------(((----....)))))))).((((.((((((..((...........))..))))))...)))).............. ( -19.90) >DroEre_CAF1 61009 90 - 1 GUGCGUACAAGUGUGU--GUGU----GCGUGUGCACAUAGUUUUGCGGCAGCUGUGUAAAUAUUUUAAAUGCCGCUUUAGACCUAAAUGCACCCAA ((((((.........(--((((----((....)))))))((((.((((((..((...........))..))))))...))))....)))))).... ( -27.20) >DroWil_CAF1 59111 88 - 1 GUGGGUGUGUGU--GU--GUGU----CUGGGUGCACAUAGUUUUGCGGCAUCUGUGUAAAUAUUUUAAACGCCCUUUUAGACUUAAAUGUACUCAA .((((((..(..--.(--(.((----(((((((.....(((((((((.(....))))))).))).....)))))....)))).)).)..)))))). ( -20.80) >DroYak_CAF1 57086 88 - 1 GUGCGUACAAGUGUGU----GU----GAGUGUGCACAUAGUUUUGCGGCAGCUGUGUAAAUAUUUUAAAUGCCGCUUUAGACCCAAAUGCACACCA ((((((.....(((((----((----......)))))))((((.((((((..((...........))..))))))...))))....)))))).... ( -27.00) >DroPer_CAF1 54894 78 - 1 GUGUA--------------UAU----UUGUGUGCACAUAGUUUUGCGGCAGCUGUGUAAAUAUUUUAAAUGCCGCUUUAGACCUAAAUGCACCCUA (((((--------------((.----...)))))))...((((.((((((..((...........))..))))))...)))).............. ( -18.70) >consensus GUGCGUACAAGU__GU___UGU____CUGUGUGCACAUAGUUUUGCGGCAGCUGUGUAAAUAUUUUAAAUGCCGCUUUAGACCUAAAUGCACCCAA ..............................(((((....((((.((((((..((...........))..))))))...)))).....))))).... (-15.86 = -15.92 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:25 2006