
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,374,219 – 9,374,346 |
| Length | 127 |
| Max. P | 0.934603 |

| Location | 9,374,219 – 9,374,319 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -19.18 |
| Energy contribution | -18.93 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
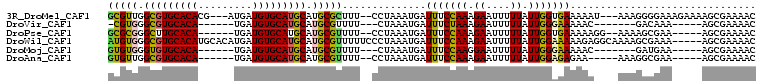
>3R_DroMel_CAF1 9374219 100 - 27905053 GCGUUGGCGUGCACACG---AUGAUGUGCAUGCAUGCGCUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAU---AAAGGGGAAAGAAAAGCGAAAAC ((((..(((((((((..---....)))))))))..)).((((--(((.....((((((((.((....)).))))....))))---....)))))))....))...... ( -28.00) >DroVir_CAF1 60650 86 - 1 -CGUGGGCGUGCACA------UGAUGUGCAUGCAUGCGUUUU---CUAAAUGAUUUCUAAAGAAUUUUUAUUGGGAAAAAC-------GACAAA-----AGCGAAAAC -(((..((((((((.------....)))))))).((((((((---((.(((((.(((....)))...))))).)).)))))-------).))..-----.)))..... ( -25.40) >DroPse_CAF1 8719 93 - 1 GCGCGGGCUUGCACA------UGAUGUGCAUGCAUGCGUUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAAAGG--AAAAGCGAA-----AGCGAAAAC (((((.((.(((((.------....))))).)).)))))(((--(((.........((((.((....)).))))......)))--))).((...-----.))...... ( -27.66) >DroWil_CAF1 2590 103 - 1 AUGUGGGCGUGCACAUGCACAUGAUGUGCAUGCAUGCGUUUUUCCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGAAAAAGAGGCAAAAGCGAAA-----AGCGAAAAC ......(((((((..(((((.....))))))))))))(((((((.........(((((((.((....)).))))))).....((....))))))-----)))...... ( -30.80) >DroMoj_CAF1 67806 87 - 1 GUGUGGGUGUGCACA------UGAUGUGCAUGCAUGCGUUUU---CUAAAUGAUUUCCAAGGAAUUUUUAUUGGGAAAAAC-------GAUGAA-----AGCGAAAAC (((((.(((..((..------...))..))).)))))(((((---(.......(((((((..........)))))))....-------...)))-----)))...... ( -24.44) >DroAna_CAF1 3274 90 - 1 GUGUUGGCGUGCACA------UGAUGUGCAUGCAUGCGUUUU--CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGAGAGAA-----AAAGGCGAA-----AGCGAAAAC ..((..((((((((.------....))))))))..))(((((--(........(((((((.((....)).)))))))...-----....((...-----.)))))))) ( -28.70) >consensus GCGUGGGCGUGCACA______UGAUGUGCAUGCAUGCGUUUU__CCUAAAUGAUUUCCAAAGAAUUUUUAUUGGAAAAAAA____AAAGGCGAA_____AGCGAAAAC (((((.(((((((((.........))))))))).)))))..............(((((((.((....)).)))))))............................... (-19.18 = -18.93 + -0.25)

| Location | 9,374,244 – 9,374,346 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -17.09 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.85 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9374244 102 + 27905053 UUUUCACCAAUAAAAAUUCUUUGGAAAUCAUUUAGGAAAGCGCAUGCAUGCACAUCAUCGUGUGCACGCCAACGCCC------ACGCCC-CUAC-----UGUUCCG-CCCCGCCC ......................((((........((...(((...((.(((((((....))))))).))...)))..------...)).-....-----..)))).-........ ( -20.26) >DroPse_CAF1 8740 101 + 1 UUUUCACCAAUAAAAAUUCUUUGGAAAUCAUUUAGGAAAACGCAUGCAUGCACAUCA---UGUGCAAGCCCGCGCCC------ACACCCGCUAC-----CAGACAGACCCCUUCU ......((((..........))))..........((.....((..((.(((((....---.))))).))..))((..------......))..)-----)............... ( -16.90) >DroEre_CAF1 4581 107 + 1 UUUUCACCAAUAAAAAUUCUUGGGAAAUCAUUUAGGAAAGCGCAUGCAUGCACAUCAUCAUGUGCACGCCAACGCCCACGCCCACGCCC-CUAC-----CGUUUCG-CCCCGCC- ......((((.........))))(((((....((((...(((...((.(((((((....))))))).))...)))....((....)).)-))).-----.))))).-.......- ( -22.80) >DroMoj_CAF1 67822 91 + 1 UUUUUCCCAAUAAAAAUUCCUUGGAAAUCAUUUAG-AAAACGCAUGCAUGCACAUCA---UGUGCACACCCACACAC------ACACAC-A--------CAGAAAGAGCG----- ((((((((((..........))))...........-............(((((....---.)))))...........------......-.--------..))))))...----- ( -10.60) >DroAna_CAF1 3292 102 + 1 UUCUCUCCAAUAAAAAUUCUUUGGAAAUCAUUUAGGAAAACGCAUGCAUGCACAUCA---UGUGCACGCCAACACCC------ACGCUC-CAAGCCUGCCCCUACC-AUCCAC-- .....(((((..........))))).......((((.........((.(((((....---.))))).)).......(------(.((..-...)).))..))))..-......-- ( -17.70) >DroPer_CAF1 8681 91 + 1 UUUUCACCAAUAAAAAUUCUUUGGAAAUCAUUUAGGAAAACGCAUGCAUGCACAUCA---UGUGCAAGCCCACGCCC------ACACCCGCUCC-----CCGACA---------- ......((((..........))))..........(((........((.(((((....---.))))).))....((..------......)))))-----......---------- ( -14.30) >consensus UUUUCACCAAUAAAAAUUCUUUGGAAAUCAUUUAGGAAAACGCAUGCAUGCACAUCA___UGUGCACGCCAACGCCC______ACACCC_CUAC_____CAGACAG_CCCC_C__ ......((((..........)))).....................((.((((((......)))))).)).............................................. (-10.52 = -10.85 + 0.33)



| Location | 9,374,244 – 9,374,346 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -20.14 |
| Energy contribution | -19.78 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9374244 102 - 27905053 GGGCGGGG-CGGAACA-----GUAG-GGGCGU------GGGCGUUGGCGUGCACACGAUGAUGUGCAUGCAUGCGCUUUCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAA ........-.((((..-----((..-(((...------((((((..(((((((((......)))))))))..)))))).)))..))...))))...................... ( -30.10) >DroPse_CAF1 8740 101 - 1 AGAAGGGGUCUGUCUG-----GUAGCGGGUGU------GGGCGCGGGCUUGCACA---UGAUGUGCAUGCAUGCGUUUUCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAA ....(..(((.((...-----...))(((...------(((((((.((.(((((.---....))))).)).))))))).)))....)))..)....................... ( -25.70) >DroEre_CAF1 4581 107 - 1 -GGCGGGG-CGAAACG-----GUAG-GGGCGUGGGCGUGGGCGUUGGCGUGCACAUGAUGAUGUGCAUGCAUGCGCUUUCCUAAAUGAUUUCCCAAGAAUUUUUAUUGGUGAAAA -...((((-((..(((-----....-...)))...)))((((((..((((((((((....))))))))))..)))))).)))......((((((((.........)))).)))). ( -35.80) >DroMoj_CAF1 67822 91 - 1 -----CGCUCUUUCUG--------U-GUGUGU------GUGUGUGGGUGUGCACA---UGAUGUGCAUGCAUGCGUUUU-CUAAAUGAUUUCCAAGGAAUUUUUAUUGGGAAAAA -----(((......((--------(-(((..(------(((((((......))))---).)))..)))))).)))((((-((.(((((.(((....)))...))))).)))))). ( -23.30) >DroAna_CAF1 3292 102 - 1 --GUGGAU-GGUAGGGGCAGGCUUG-GAGCGU------GGGUGUUGGCGUGCACA---UGAUGUGCAUGCAUGCGUUUUCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGAGAGAA --......-..((((((((.((...-..)).)------)...((..((((((((.---....))))))))..))...)))))).....(((((((.((....)).)))))))... ( -28.80) >DroPer_CAF1 8681 91 - 1 ----------UGUCGG-----GGAGCGGGUGU------GGGCGUGGGCUUGCACA---UGAUGUGCAUGCAUGCGUUUUCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAA ----------....((-----..((((.((((------((((....))))((((.---....))))..)))).))))..)).......((.((((.((....)).)))).))... ( -26.00) >consensus __G_GGGG_CGGACCG_____GUAG_GGGCGU______GGGCGUGGGCGUGCACA___UGAUGUGCAUGCAUGCGUUUUCCUAAAUGAUUUCCAAAGAAUUUUUAUUGGUGAAAA ........................................((((..(((((((((......)))))))))..))))((((((((..((.(((....))).))...)))).)))). (-20.14 = -19.78 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:54 2006