
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,368,370 – 9,368,597 |
| Length | 227 |
| Max. P | 0.999622 |

| Location | 9,368,370 – 9,368,469 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368370 99 - 27905053 AGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAAAAGGCGAAACAAAAGCAAGACAAUCCAUGGGCCAUAUACA (((....)))(((((((((((((.(((.......))))))).))))...))))).......(((.........................)))....... ( -23.51) >DroSec_CAF1 137693 99 - 1 AGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAAAAGGCGAAACAAAAGCAAGACUAUAUAUACGCUAUAUACG (((....)))(((((((((((((.(((.......))))))).))))...))))).......((((.......((.....)).......))))....... ( -23.24) >DroSim_CAF1 146795 99 - 1 AGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAAAAGGCGAAACAAAAGCAAGACAAUCCAUGGGCUAUAUACA (((...(((((((((((((((((.(((.......))))))).))))...))))).......(......)...)))).......(....))))....... ( -23.20) >DroEre_CAF1 138405 99 - 1 AGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAAAAGGCGAAACAAACGCAAGACAAACCAUGGGCUAUAUACA .(((.....)))(((....((((.(((.......)))))))((((((....((((......))))....))))))..............)))....... ( -24.70) >DroYak_CAF1 138872 99 - 1 AGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAAAAGGCGAAACAAAAGCAAGACAAACCAUGGGCUAUAUACA (((...(((((((((((((((((.(((.......))))))).))))...))))).......(......)...)))).......(....))))....... ( -23.60) >consensus AGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAAAAGGCGAAACAAAAGCAAGACAAUCCAUGGGCUAUAUACA (((....)))(((((((((((((.(((.......))))))).))))...)))(((......))).........))........................ (-21.92 = -21.92 + -0.00)

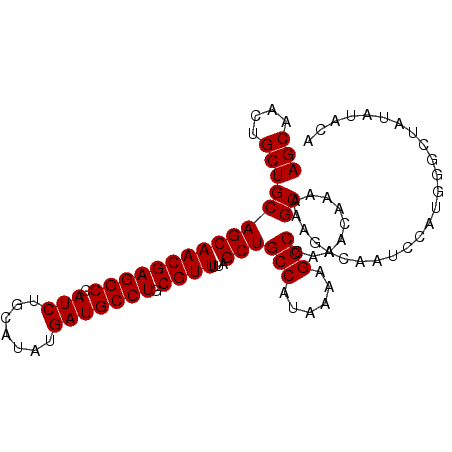

| Location | 9,368,410 – 9,368,505 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -34.58 |
| Energy contribution | -34.58 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368410 95 + 27905053 UUAUGGCAGCUAAAACGCAGGCAUCAUAUGCAGAUGGCCUCGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUG ..((((((((((.((((.(((((((.......))).))))))))..(((((((((....)))))))))......))))))))))........... ( -34.60) >DroSec_CAF1 137733 95 + 1 UUAUGGCAGCUAAAACGCAGGCAUCAUAUGCAGAUGGCCUCGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUG ..((((((((((.((((.(((((((.......))).))))))))..(((((((((....)))))))))......))))))))))........... ( -34.60) >DroSim_CAF1 146835 95 + 1 UUAUGGCAGCUAAAACGCAGGCAUCAUAUGCAGAUGGCCUCGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUG ..((((((((((.((((.(((((((.......))).))))))))..(((((((((....)))))))))......))))))))))........... ( -34.60) >DroEre_CAF1 138445 95 + 1 UUAUGGCAGCUAAAACGCAGGCAUCAUAUGCAGAUGGCCUCGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUG ..((((((((((.((((.(((((((.......))).))))))))..(((((((((....)))))))))......))))))))))........... ( -34.60) >DroYak_CAF1 138912 95 + 1 UUAUGGCAGCUAAAACGCAGGCAUCAUAUGCAGAUGGCCUCGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUGUAGUUGCUGUUCGCGCAUCUG ..((((((((((.((((.(((((((.......))).))))))))..(((((((((....)))))))))......))))))))))........... ( -34.50) >consensus UUAUGGCAGCUAAAACGCAGGCAUCAUAUGCAGAUGGCCUCGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUG ..((((((((((.((((.(((((((.......))).))))))))..(((((((((....)))))))))......))))))))))........... (-34.58 = -34.58 + -0.00)



| Location | 9,368,410 – 9,368,505 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368410 95 - 27905053 CAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAA (((.(((((.....(((.((....)).)))....)).))).)))..(((((((((((((.(((.......))))))).))))...)))))..... ( -29.30) >DroSec_CAF1 137733 95 - 1 CAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAA (((.(((((.....(((.((....)).)))....)).))).)))..(((((((((((((.(((.......))))))).))))...)))))..... ( -29.30) >DroSim_CAF1 146835 95 - 1 CAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAA (((.(((((.....(((.((....)).)))....)).))).)))..(((((((((((((.(((.......))))))).))))...)))))..... ( -29.30) >DroEre_CAF1 138445 95 - 1 CAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAA (((.(((((.....(((.((....)).)))....)).))).)))..(((((((((((((.(((.......))))))).))))...)))))..... ( -29.30) >DroYak_CAF1 138912 95 - 1 CAGAUGCGCGAACAGCAACUACACAGAUGCAACAGCAGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAA (((.(((((.....(((.((....)).)))....)).))).)))..(((((((((((((.(((.......))))))).))))...)))))..... ( -29.30) >consensus CAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACGAGGCCAUCUGCAUAUGAUGCCUGCGUUUUAGCUGCCAUAA (((.(((((.....(((.((....)).)))....)).))).)))..(((((((((((((.(((.......))))))).))))...)))))..... (-29.30 = -29.30 + 0.00)

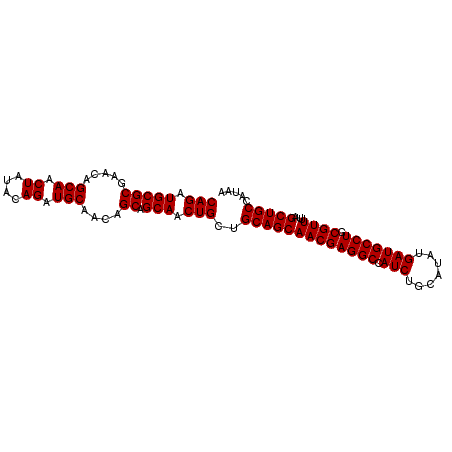

| Location | 9,368,450 – 9,368,545 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368450 95 + 27905053 CGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAAC .((((((((((((((....))))))))).......((.(((.......))).)))))))...(((((((((...........))))...))))). ( -26.70) >DroSec_CAF1 137773 95 + 1 CGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAAC .((((((((((((((....))))))))).......((.(((.......))).)))))))...(((((((((...........))))...))))). ( -26.70) >DroSim_CAF1 146875 95 + 1 CGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAAC .((((((((((((((....))))))))).......((.(((.......))).)))))))...(((((((((...........))))...))))). ( -26.70) >DroEre_CAF1 138485 95 + 1 CGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAAC .((((((((((((((....))))))))).......((.(((.......))).)))))))...(((((((((...........))))...))))). ( -26.70) >DroYak_CAF1 138952 95 + 1 CGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUGUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAAC .((((((((((((((....)))))))))..((((..(.......)..))))...)))))...(((((((((...........))))...))))). ( -27.10) >consensus CGUUGCUGCAGCAGUUGCUGCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAAC .((((((((((((((....))))))))).......((.(((.......))).)))))))...(((((((((...........))))...))))). (-26.70 = -26.70 + -0.00)



| Location | 9,368,450 – 9,368,545 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368450 95 - 27905053 GUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACG ((((((.....((((..((((.((((((.....)).))))))))(((.......))).........))))....(((((....))))))))))). ( -27.60) >DroSec_CAF1 137773 95 - 1 GUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACG ((((((.....((((..((((.((((((.....)).))))))))(((.......))).........))))....(((((....))))))))))). ( -27.60) >DroSim_CAF1 146875 95 - 1 GUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACG ((((((.....((((..((((.((((((.....)).))))))))(((.......))).........))))....(((((....))))))))))). ( -27.60) >DroEre_CAF1 138485 95 - 1 GUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACG ((((((.....((((..((((.((((((.....)).))))))))(((.......))).........))))....(((((....))))))))))). ( -27.60) >DroYak_CAF1 138952 95 - 1 GUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUACACAGAUGCAACAGCAGCAACUGCUGCAGCAACG ((((((.....((((..((((.((((((.....)).))))))))(((.......))).........))))....(((((....))))))))))). ( -27.60) >consensus GUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGCAGCAACUGCUGCAGCAACG ((((((.....((((..((((.((((((.....)).))))))))(((.......))).........))))....(((((....))))))))))). (-27.60 = -27.60 + -0.00)



| Location | 9,368,469 – 9,368,575 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368469 106 + 27905053 GCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGC ((....))....((((.((((.((((.((((...(((((((...((((((((...........)))......)))))..)))))))..)).)))))).)))))))) ( -28.20) >DroSec_CAF1 137792 106 + 1 GCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGC ((....))....((((.((((.((((.((((...(((((((...((((((((...........)))......)))))..)))))))..)).)))))).)))))))) ( -28.20) >DroSim_CAF1 146894 106 + 1 GCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGC ((....))....((((.((((.((((.((((...(((((((...((((((((...........)))......)))))..)))))))..)).)))))).)))))))) ( -28.20) >consensus GCUGUUGCAUCUGUAUAGUUGCUGUUCGCGCAUCUGCAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGC ((....))....((((.((((.((((.((((...(((((((...((((((((...........)))......)))))..)))))))..)).)))))).)))))))) (-28.20 = -28.20 + -0.00)



| Location | 9,368,469 – 9,368,575 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -32.60 |
| Energy contribution | -32.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368469 106 - 27905053 GCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGC ((((.((((.(((((((..(((((....(((((.......)))))..((((.((((((.....)).)))))))))))))...)))))))...))))))))...... ( -32.60) >DroSec_CAF1 137792 106 - 1 GCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGC ((((.((((.(((((((..(((((....(((((.......)))))..((((.((((((.....)).)))))))))))))...)))))))...))))))))...... ( -32.60) >DroSim_CAF1 146894 106 - 1 GCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGC ((((.((((.(((((((..(((((....(((((.......)))))..((((.((((((.....)).)))))))))))))...)))))))...))))))))...... ( -32.60) >consensus GCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUGCAGAUGCGCGAACAGCAACUAUACAGAUGCAACAGC ((((.((((.(((((((..(((((....(((((.......)))))..((((.((((((.....)).)))))))))))))...)))))))...))))))))...... (-32.60 = -32.60 + 0.00)


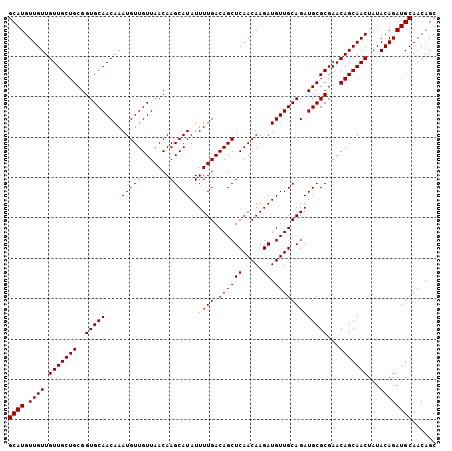
| Location | 9,368,505 – 9,368,597 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368505 92 + 27905053 CAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGCGGCAAGAGGCAUGCGGCAUGCA .......((((((.((((((((...(((.(((....))))))...)))....)))))..))))))(((((.(((.......))).))))).. ( -25.00) >DroSec_CAF1 137828 92 + 1 CAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGCGGCAAGAGGCAUGCGGCAUGCA .......((((((.((((((((...(((.(((....))))))...)))....)))))..))))))(((((.(((.......))).))))).. ( -25.00) >DroSim_CAF1 146930 92 + 1 CAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGCGGCAAGAGGCAUGCGGCAUGCA .......((((((.((((((((...(((.(((....))))))...)))....)))))..))))))(((((.(((.......))).))))).. ( -25.00) >consensus CAACAUCUUGUUGAGCUGUCAAAAUAUGCUUGUUAACAACAUUUGUUGCACCGCAGCAACAACAACAUGCGGCAAGAGGCAUGCGGCAUGCA .......((((((.((((((((...(((.(((....))))))...)))....)))))..))))))(((((.(((.......))).))))).. (-25.00 = -25.00 + 0.00)


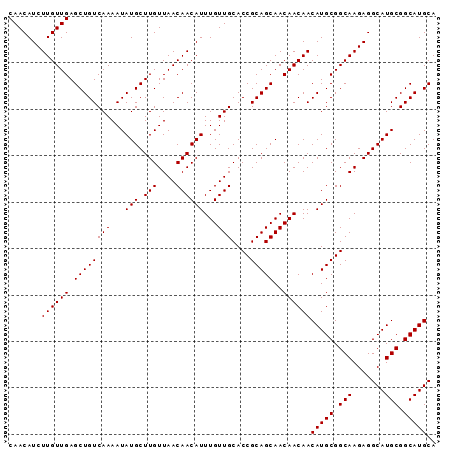
| Location | 9,368,505 – 9,368,597 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9368505 92 - 27905053 UGCAUGCCGCAUGCCUCUUGCCGCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUG .((((((.(((.......))).))))))..(((((((......)))))))..((((((((..........)))))))).((((.....)))) ( -27.00) >DroSec_CAF1 137828 92 - 1 UGCAUGCCGCAUGCCUCUUGCCGCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUG .((((((.(((.......))).))))))..(((((((......)))))))..((((((((..........)))))))).((((.....)))) ( -27.00) >DroSim_CAF1 146930 92 - 1 UGCAUGCCGCAUGCCUCUUGCCGCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUG .((((((.(((.......))).))))))..(((((((......)))))))..((((((((..........)))))))).((((.....)))) ( -27.00) >consensus UGCAUGCCGCAUGCCUCUUGCCGCAUGUUGUUGUUGCUGCGGUGCAACAAAUGUUGUUAACAAGCAUAUUUUGACAGCUCAACAAGAUGUUG .((((((.(((.......))).))))))..(((((((......)))))))..((((((((..........)))))))).((((.....)))) (-27.00 = -27.00 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:51 2006