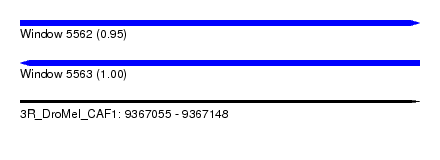
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,367,055 – 9,367,148 |
| Length | 93 |
| Max. P | 0.998437 |

| Location | 9,367,055 – 9,367,148 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.24 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9367055 93 + 27905053 UUUUGGGCUUUUUUUUUUGUUGUUGUCCGGUUGGUUGCAUUUUGUGCAUGUCAAGCCAAGGUUCAUUAAGCUGAAUUUUUCAGCAAUUUGGAA ....((((................))))((((...(((((...))))).....))))....((((....(((((.....)))))....)))). ( -20.09) >DroSec_CAF1 136398 91 + 1 UUUUGGGCUUUUUUUUU--UUGUUGUCCGGCUGGCUGCAUUUUGUGCAUGUCAAGCCAAGGUUCAUUAAGCUGAAUUUUUCAGCAAUUUGCAA ....((((.........--.....))))((((((((((((...))))).))).))))............(((((.....)))))......... ( -24.94) >DroSim_CAF1 145502 90 + 1 UUUUGGGCUUUUU-UUU--UUGUUGUCCGGCUGGCUGCAUUUUGUGCAUGUCAAGCCAAGGUUCAUUAAGCUGAAUUUUUCAGCAAUUUGCAA ....((((.....-...--.....))))((((((((((((...))))).))).))))............(((((.....)))))......... ( -25.02) >DroEre_CAF1 137089 93 + 1 UUUAAGUUUUUUUUUCUUUUUGUUGCUCGGCUGACUGCAUUUUGUGCAUGUCAAGCCAAGGUUCAUUAAGCUAAAUUUUUCAGCAAUUUGCAA .....................((((((.((((((((((((...))))).))).))))..((((.....)))).........))))))...... ( -20.10) >DroYak_CAF1 137556 90 + 1 UUUUGUUUU--UUUUUUUUUUGUU-UUCAGCUGGCUGCAUUUUGUGCAUGUCAAGCCAAGGUUCAUUAAGCUAAAUUUUUCAGCAAUUUGCAA .........--.............-...((((((((((((...))))).))).(((....))).....))))..........((.....)).. ( -12.60) >consensus UUUUGGGCUUUUUUUUUU_UUGUUGUCCGGCUGGCUGCAUUUUGUGCAUGUCAAGCCAAGGUUCAUUAAGCUGAAUUUUUCAGCAAUUUGCAA ...................((((((...((((((((((((...))))).))).))))((((((((......)))))))).))))))....... (-16.92 = -17.24 + 0.32)

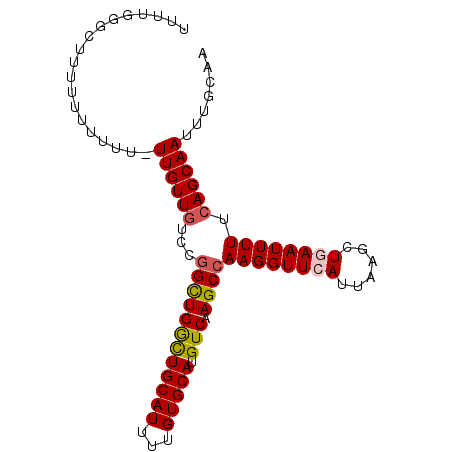
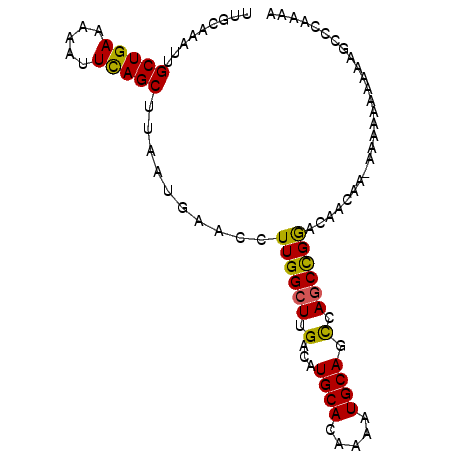
| Location | 9,367,055 – 9,367,148 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -17.72 |
| Consensus MFE | -14.46 |
| Energy contribution | -13.86 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9367055 93 - 27905053 UUCCAAAUUGCUGAAAAAUUCAGCUUAAUGAACCUUGGCUUGACAUGCACAAAAUGCAACCAACCGGACAACAACAAAAAAAAAAGCCCAAAA .........(((((.....)))))............(((((....((((.....)))).((....))................)))))..... ( -15.80) >DroSec_CAF1 136398 91 - 1 UUGCAAAUUGCUGAAAAAUUCAGCUUAAUGAACCUUGGCUUGACAUGCACAAAAUGCAGCCAGCCGGACAACAA--AAAAAAAAAGCCCAAAA ..((.....(((((.....)))))..........((((((.(...((((.....)))).).)))))).......--.........))...... ( -18.30) >DroSim_CAF1 145502 90 - 1 UUGCAAAUUGCUGAAAAAUUCAGCUUAAUGAACCUUGGCUUGACAUGCACAAAAUGCAGCCAGCCGGACAACAA--AAA-AAAAAGCCCAAAA ..((.....(((((.....)))))..........((((((.(...((((.....)))).).)))))).......--...-.....))...... ( -18.30) >DroEre_CAF1 137089 93 - 1 UUGCAAAUUGCUGAAAAAUUUAGCUUAAUGAACCUUGGCUUGACAUGCACAAAAUGCAGUCAGCCGAGCAACAAAAAGAAAAAAAAACUUAAA ((((.....(((((.....)))))..........((((((.(((.((((.....)))))))))))))))))...................... ( -22.60) >DroYak_CAF1 137556 90 - 1 UUGCAAAUUGCUGAAAAAUUUAGCUUAAUGAACCUUGGCUUGACAUGCACAAAAUGCAGCCAGCUGAA-AACAAAAAAAAAA--AAAACAAAA ..((.....(((((.....)))))...........(((((.(.(((.......)))))))))))....-.............--......... ( -13.60) >consensus UUGCAAAUUGCUGAAAAAUUCAGCUUAAUGAACCUUGGCUUGACAUGCACAAAAUGCAGCCAGCCGGACAACAA_AAAAAAAAAAGCCCAAAA .........(((((.....)))))..........((((((.(...((((.....)))).).)))))).......................... (-14.46 = -13.86 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:43 2006