
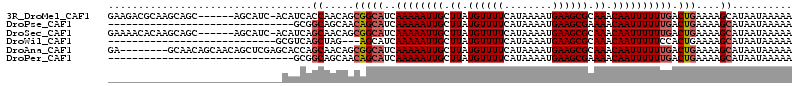
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,361,233 – 9,361,365 |
| Length | 132 |
| Max. P | 0.581553 |

| Location | 9,361,233 – 9,361,340 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -15.57 |
| Consensus MFE | -8.73 |
| Energy contribution | -9.15 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9361233 107 + 27905053 GAAGACGCAAGCAGC------AGCAUC-ACAUCACCAACAGCGGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAA (((((((.(((((((------.((...-............)).)).........))))).)))))))..........((.((..(((....)))..))....)).......... ( -15.76) >DroPse_CAF1 126062 83 + 1 -------------------------------GCGGCAGCAACAGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGAAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAA -------------------------------((..((((((.((((........))))..(((((((............))))))).....))).)))....)).......... ( -14.50) >DroSec_CAF1 130842 107 + 1 GAAAACACAAGCAGC------AGCAUC-ACAUCAGCAACAGCGGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAA (((((((.((((((.------......-......((....))...........)))))).)))))))..........((.((..(((....)))..))....)).......... ( -16.55) >DroWil_CAF1 131219 83 + 1 ----------------------------GCGUCAGCUAG---AGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUCCACUGAAAAGCAUAAUAAAAA ----------------------------((.((((...(---((((........))))).(((((((((...)))))).))).............))))...)).......... ( -14.50) >DroAna_CAF1 125964 106 + 1 GA--------GCAACAGCAACAGCUCGAGCACCAGCAACAGCGGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAA ((--------((..........))))..((.((.((....))))((((((((((((.((.((((((........)))))).)).))).))))))).))....)).......... ( -17.60) >DroPer_CAF1 131638 83 + 1 -------------------------------GCGGCAGCAACAGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGAAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAA -------------------------------((..((((((.((((........))))..(((((((............))))))).....))).)))....)).......... ( -14.50) >consensus _____________________________CAUCAGCAACAGCAGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAA ..................................((.....(((((..((((((((.((.((((((........)))))).)).)))))))))).)))....)).......... ( -8.73 = -9.15 + 0.42)


| Location | 9,361,260 – 9,361,365 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -12.38 |
| Consensus MFE | -8.63 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9361260 105 + 27905053 CCAACAGCGGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAACAACAA--------AAUUCCA----CACACAAAAACA--- ......((..((((((((((((.((.((((((........)))))).)).))).))))))).))....))................--------.......----............--- ( -11.50) >DroPse_CAF1 126065 114 + 1 GCAGCAACAGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGAAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAAGGCAGAAACAAAAACAAGACA----GA--CAAAAAGAGAG ((((((..((((........))))..))))(((((...)))))))........(((((((.(((....((...........)).....(........).))----).--))))))).... ( -12.90) >DroWil_CAF1 131225 102 + 1 GCUAG---AGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUCCACUGAAAAGCAUAAUAAAAACAAAAA--------AAAUACA--ACAA--CAGCAACA--- (((.(---((((........))))).(((((((((...)))))).))).....((((((....)))))).................--------.......--....--.)))....--- ( -11.60) >DroYak_CAF1 131516 103 + 1 GCAACAGCGGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAACAACAA--------AAUUCCG----CA--CAAAAACA--- ......((((....((((((((.((.((((((........)))))).)).))))))))((.((....))))...............--------....)))----).--........--- ( -15.10) >DroAna_CAF1 125990 105 + 1 GCAACAGCGGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAACAAAAA----------UACAGAAACAA--CAACAACA--- ((.....(((((..((((((((.((.((((((........)))))).)).)))))))))).)))....))................----------...........--........--- ( -11.60) >DroPer_CAF1 131641 113 + 1 GCAGCAACAGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGAAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAAGACAGAAACAAAAACAAGACA----GA--CAAAAAGAGA- ((((((..((((........))))..))))(((((...)))))))........(((((((.(((...................................))----).--)))))))...- ( -11.55) >consensus GCAACAGCAGCAUCAAAAAUUGCUUAUGUUUUCAUAAAAUGAAGCGCAAACAAUUUUUUGACUGAAAAGCAUAAUAAAAACAAAAA________AAAUACA____AA__CAAAAACA___ ((.....(((((..((((((((.((.((((((........)))))).)).)))))))))).)))....)).................................................. ( -8.63 = -9.05 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:34 2006