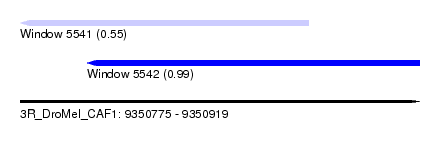
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,350,775 – 9,350,919 |
| Length | 144 |
| Max. P | 0.986189 |

| Location | 9,350,775 – 9,350,879 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -14.92 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
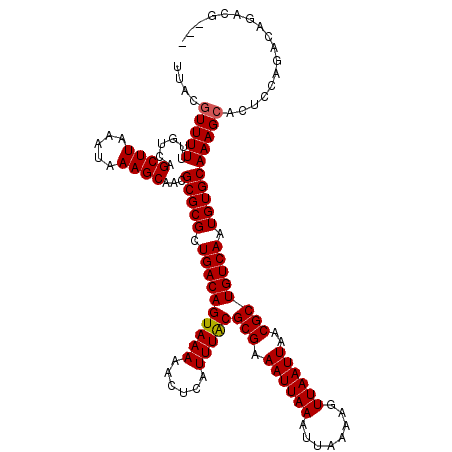
>3R_DroMel_CAF1 9350775 104 - 27905053 CAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGUCAUUCCUCCUCCUUACACCUCG--CCCUCUCCUUA ..(((((......)))))((((((((((........))))))...((((.....((((...))))..))))...................)))--).......... ( -13.10) >DroSec_CAF1 120199 104 - 1 CAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGCCAUUCCUCCUGCUUCCACCUCG--CCCUCUCCUUU ..(((((......)))))((((((((((........))))))...((((.....((((...))))..))))...................)))--).......... ( -15.20) >DroEre_CAF1 120568 104 - 1 CAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGCCAUUCGCCCUCCUUCCACCUCA--CCCUCUUCACU ..(((((......)))))(((((............(((....)))((((.....((((...))))..))))..)))))...............--........... ( -15.90) >DroYak_CAF1 121643 106 - 1 CAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGCCAUUCGUCCUCCUUCCACCUCCACCCCUCUUCACU ..(((((......)))))(((((............(((....)))((((.....((((...))))..))))..)))))............................ ( -13.80) >DroAna_CAF1 116075 84 - 1 CAGUAAAAACUCAUUUGCGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGACAGCAGACUUU---AC------------------- ..(((((......)))))....((((((........))))))...((((((...((((...))))....)))))).......---..------------------- ( -16.60) >consensus CAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGCCAUUCGUCCUCCUUCCACCUCG__CCCUCUCCACU ..(((((......)))))(((.((((((........))))))..))).......((((...))))......................................... ( -9.36 = -9.20 + -0.16)

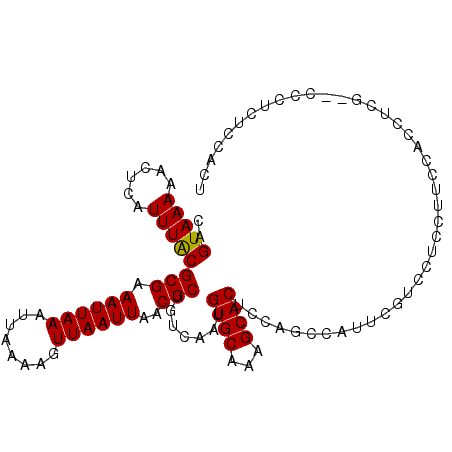
| Location | 9,350,799 – 9,350,919 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -24.09 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9350799 120 - 27905053 UUGCGUUUUUUGUCAGCUUAAAUAAAGCAACGCGCGCUGACAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGUCAUUCCUCC .(((...........((((.....))))...(((((.((((((((((......)))))(((.((((((........))))))..)))))))).)))))...)))................ ( -25.80) >DroPse_CAF1 115217 117 - 1 UUACGUUUUUUGUCAGCUUAAAUAAAGCAACGCGCGCUGACAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGACAGACU--- ........((((((.((((.....))))...(((((.((((((((((......)))))(((.((((((........))))))..)))))))).)))))...........))))))..--- ( -28.90) >DroEre_CAF1 120592 120 - 1 UUGCGUUUUUUGUCAGCUUAAAUAAAGCAACGCGCGCUGACAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGCCAUUCGCCC .((.(((((((((((((.........((.....)))))))))..)))))).)).....(((((............(((....)))((((.....((((...))))..))))..))))).. ( -27.00) >DroWil_CAF1 118927 114 - 1 UUGC-UUUUUUGUCAGCUUAAAUAAAGCAAUGCGCGCUGACAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUUCAGUCAGC----- .(((-((........((((.....))))..((((((.((((((((((......)))))(((.((((((........))))))..)))))))).)))))))))))...........----- ( -27.50) >DroAna_CAF1 116079 120 - 1 UUACGUUUUUUGUCAGCUUAAAUAAAGCAACGCGCGCUGACAGUAAAAACUCAUUUGCGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGACAGCAGACU ....((((.(((((.((((.....))))...(((((.((((((((((......)))))(((.((((((........))))))..)))))))).)))))...........))))).)))). ( -31.30) >DroPer_CAF1 119988 117 - 1 UUACGUUUUUUGUCAGCUUAAAUAAAGCAACGCGCGCUGACAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGACAGACU--- ........((((((.((((.....))))...(((((.((((((((((......)))))(((.((((((........))))))..)))))))).)))))...........))))))..--- ( -28.90) >consensus UUACGUUUUUUGUCAGCUUAAAUAAAGCAACGCGCGCUGACAGUAAAAACUCAUUUACGCGAAAUUAAAUUAAAAGUUAAUUAACGCUGUCAAUGUGCAAAGCACUCCAGACAGACG___ ....(((((......((((.....))))...(((((.((((((((((......)))))(((.((((((........))))))..)))))))).))))))))))................. (-24.09 = -24.12 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:23 2006