
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,349,127 – 9,349,217 |
| Length | 90 |
| Max. P | 0.618037 |

| Location | 9,349,127 – 9,349,217 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
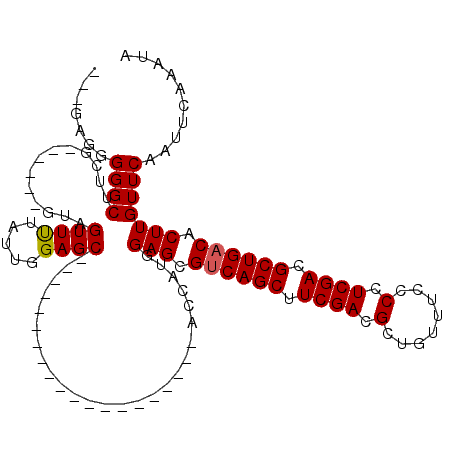
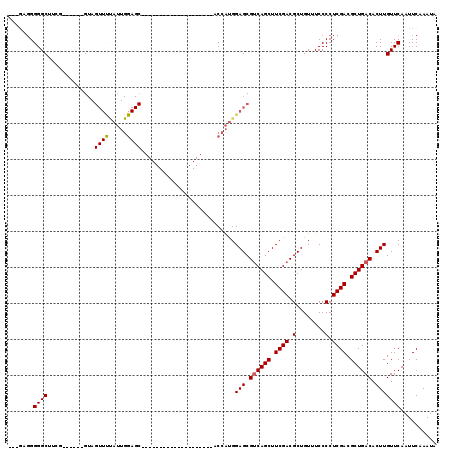
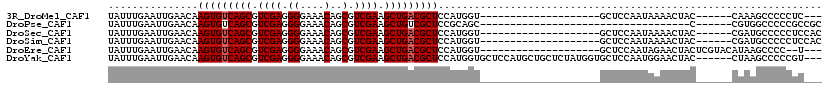
>3R_DroMel_CAF1 9349127 90 + 27905053 ---GAGGGGGCUUUG------GUAGUUUUAUUGGAGC--------------------ACCAUGGAGCGUCAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA ---((((((((((..------(((....)))..))))--------------------.......((((((......))))))....))))))((....(((....)))....))..... ( -30.80) >DroPse_CAF1 113792 78 + 1 GCGGCGGGGGCCACG------G-----------------------------------GCUGCGGAGCGACAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA (((.((((((..(((------(-----------------------------------.(..((((((....))))))..)))))..)))).)).)))...................... ( -28.60) >DroSec_CAF1 118592 93 + 1 GUGGAGGGGGCAUCG------GUAGUUUUAUUGGAGC--------------------ACCAUGGAGCGUCAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA ((.((((((((((.(------((.((((.....))))--------------------)))))).((((((......))))))...))))))).))........................ ( -31.80) >DroSim_CAF1 129847 93 + 1 GUGGAGGGGGCAUCG------GUAGUUUUAUUGGAGC--------------------ACCAUGGAGCGUCAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA ((.((((((((((.(------((.((((.....))))--------------------)))))).((((((......))))))...))))))).))........................ ( -31.80) >DroEre_CAF1 118908 94 + 1 ---A--GGGGCUUAUGUACGAGUAGUUCUAUUGGAGC--------------------ACCAUGGAGCGUCAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA ---.--........((.((((((.(((((((.((...--------------------.)))))))))((((((.((((.(........).)))).)))))))))))).))......... ( -29.70) >DroYak_CAF1 119908 110 + 1 ---ACGGGGGCUUAG------GUAGUUCCAUUGGAGCACCAUAGAGCAGCAUGGAGCACCAUGGAGCGUCAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA ---.((((((....(------((.(((((...))))))))...(((((((((((....))))(((((....)))))...))))))).)))))).......................... ( -37.50) >consensus ___GAGGGGGCUUCG______GUAGUUUUAUUGGAGC____________________ACCAUGGAGCGUCAGCUUCGACGCUGUUUCCCCUCGACGCUGACACUUGUUCAAUUCAAAUA .......((((.............((((.....))))..........................(((.((((((.((((.(........).)))).)))))).))))))).......... (-18.55 = -18.72 + 0.17)

| Location | 9,349,127 – 9,349,217 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
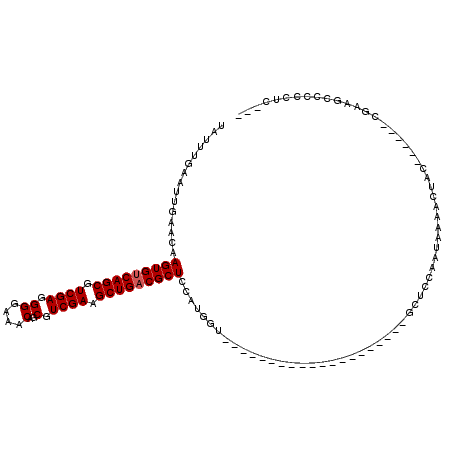
>3R_DroMel_CAF1 9349127 90 - 27905053 UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGACGCUCCAUGGU--------------------GCUCCAAUAAAACUAC------CAAAGCCCCCUC--- ..((((.........(((((((((.((((.((....)..).)))).)))))))))...(((.--------------------...)))..........------))))........--- ( -25.40) >DroPse_CAF1 113792 78 - 1 UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGUCGCUCCGCAGC-----------------------------------C------CGUGGCCCCCGCCGC ((((((.......))))))...(((.((.((((..(((((......)))))....((((...-----------------------------------.------.)))))))))).))) ( -25.50) >DroSec_CAF1 118592 93 - 1 UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGACGCUCCAUGGU--------------------GCUCCAAUAAAACUAC------CGAUGCCCCCUCCAC .......((((....(((((((((.((((.((....)..).)))).)))))))))...(((.--------------------...)))..........------))))........... ( -25.20) >DroSim_CAF1 129847 93 - 1 UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGACGCUCCAUGGU--------------------GCUCCAAUAAAACUAC------CGAUGCCCCCUCCAC .......((((....(((((((((.((((.((....)..).)))).)))))))))...(((.--------------------...)))..........------))))........... ( -25.20) >DroEre_CAF1 118908 94 - 1 UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGACGCUCCAUGGU--------------------GCUCCAAUAGAACUACUCGUACAUAAGCCCC--U--- ...............(((((((((.((((.((....)..).)))).)))))))))....((.--------------------(((......((.....)).......))).))--.--- ( -26.12) >DroYak_CAF1 119908 110 - 1 UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGACGCUCCAUGGUGCUCCAUGCUGCUCUAUGGUGCUCCAAUGGAACUAC------CUAAGCCCCCGU--- ................((((((((.((((.((....)..).)))).))))))))(((((((.((.(((((......))))).)).)).))))).....------............--- ( -38.80) >consensus UAUUUGAAUUGAACAAGUGUCAGCGUCGAGGGGAAACAGCGUCGAAGCUGACGCUCCAUGGU____________________GCUCCAAUAAAACUAC______CGAAGCCCCCUC___ ...............(((((((((.((((.((....)..).)))).)))))))))................................................................ (-21.27 = -21.43 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:21 2006