| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,327,212 – 1,327,307 |
| Length | 95 |
| Max. P | 0.999953 |

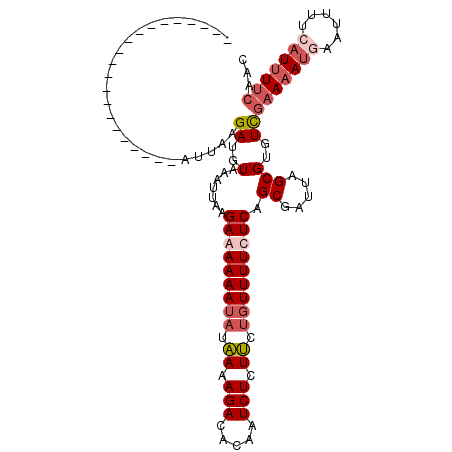
| Location | 1,327,212 – 1,327,307 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -17.17 |
| Energy contribution | -18.30 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.82 |
| SVM RNA-class probability | 0.999953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1327212 95 + 27905053 GUUGAAAAUGAAAAUUUAUUUUCAACACGCUAACCGCUGAGAAAACAGAAGAGAUUGUGUCUUUUAUAUUUUCUCUUAAUUUACAUCUUAA------------------ACAA (((((((((((....)))))))))))..((.....)).(((((((...(((((((...)))))))...)))))))................------------------.... ( -21.80) >DroSec_CAF1 15821 113 + 1 GUUGAAAAUGAAAAUUCAUUUUCGACACGCUAAUCGCUGAGAAAACAGAAGAGAUUAUGUCUUUUAUAUUUUUUCUUAAUUUACAUCUUAAUUAAGGGGGGAAUAUGAAACAA (((((((((((....)))))))))))(((.(((((.(((......)))....)))))))).(((((((((((..(((((((........)))))).)..)))))))))))... ( -30.10) >DroSim_CAF1 17520 109 + 1 GUUGAAAAUGAAAAUUCAUUUUCGACACGCUAAUCGCUGAGAAAACAGAAGAGAUUGUGUCUUUUAUAUUUUCUCUUAAUUUACAUCUUAAUUAAGAGGGGAACAUGAA---- (((((((((((....)))))))))))((((.((((.(((......)))....))))))))........((..(((((((((........)))))))))..)).......---- ( -31.80) >DroYak_CAF1 16703 84 + 1 GUAGGAA-GGAAAAU-----UUGGACACGCUAAUCGCUGAGAAAACAGCAGAGAAUGUGUCUUUAA--UUUUUUCGUAAUAUACAUCUUAAG--------------------- .(((((.-((((((.-----..(((((((....((((((......)))).))...)))))))....--.))))))(((...))).)))))..--------------------- ( -20.00) >consensus GUUGAAAAUGAAAAUUCAUUUUCGACACGCUAAUCGCUGAGAAAACAGAAGAGAUUGUGUCUUUUAUAUUUUCUCUUAAUUUACAUCUUAAU_____________________ (((((((((((....)))))))))))..((.....)).(((((((...((((((.....))))))...)))))))...................................... (-17.17 = -18.30 + 1.13)

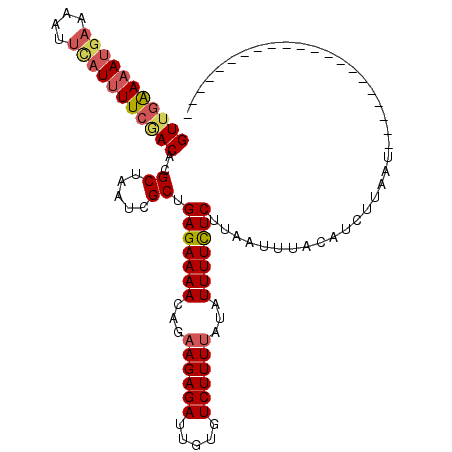
| Location | 1,327,212 – 1,327,307 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -11.62 |
| Energy contribution | -13.62 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1327212 95 - 27905053 UUGU------------------UUAAGAUGUAAAUUAAGAGAAAAUAUAAAAGACACAAUCUCUUCUGUUUUCUCAGCGGUUAGCGUGUUGAAAAUAAAUUUUCAUUUUCAAC ....------------------.....((((.((((..(((((((((.((.(((.....))).)).)))))))))...)))).))))(((((((((........))))))))) ( -18.60) >DroSec_CAF1 15821 113 - 1 UUGUUUCAUAUUCCCCCCUUAAUUAAGAUGUAAAUUAAGAAAAAAUAUAAAAGACAUAAUCUCUUCUGUUUUCUCAGCGAUUAGCGUGUCGAAAAUGAAUUUUCAUUUUCAAC .................(((((((........))))))).............(((((((((....(((......))).))))...)))))((((((((....))))))))... ( -18.10) >DroSim_CAF1 17520 109 - 1 ----UUCAUGUUCCCCUCUUAAUUAAGAUGUAAAUUAAGAGAAAAUAUAAAAGACACAAUCUCUUCUGUUUUCUCAGCGAUUAGCGUGUCGAAAAUGAAUUUUCAUUUUCAAC ----...(((((...(((((((((........)))))))))..)))))....(((((.........((......))((.....)))))))((((((((....))))))))... ( -24.60) >DroYak_CAF1 16703 84 - 1 ---------------------CUUAAGAUGUAUAUUACGAAAAAA--UUAAAGACACAUUCUCUGCUGUUUUCUCAGCGAUUAGCGUGUCCAA-----AUUUUCC-UUCCUAC ---------------------.................(((((((--((...(((((.(..((.((((......))))))..)..)))))..)-----)))))..-))).... ( -12.90) >consensus _____________________AUUAAGAUGUAAAUUAAGAAAAAAUAUAAAAGACACAAUCUCUUCUGUUUUCUCAGCGAUUAGCGUGUCGAAAAUGAAUUUUCAUUUUCAAC ..........................((..(.......(((((((((.((.(((.....))).)).))))))))).((.....)))..))(((((((......)))))))... (-11.62 = -13.62 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:54 2006