
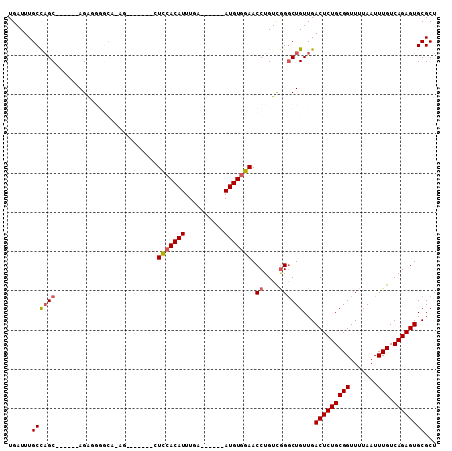
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,336,684 – 9,336,781 |
| Length | 97 |
| Max. P | 0.900333 |

| Location | 9,336,684 – 9,336,781 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -17.81 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
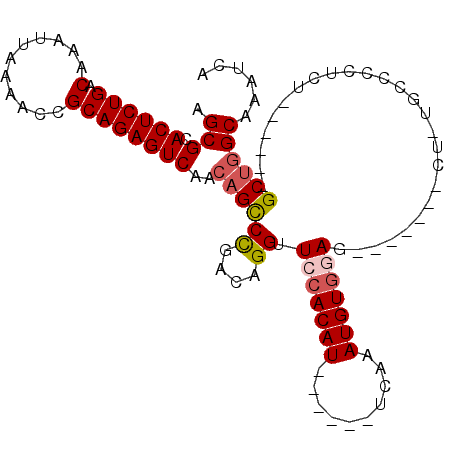
>3R_DroMel_CAF1 9336684 97 + 27905053 UGAUUUGCCAGCUCGAACAGAGG-----AG-------CUCCACAUUUGAAUGUGAAUGUGGAACCUGUUGGGCUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ......((((((((.(((((.(.-----..-------.(((((((((......))))))))).))))))))))))...(((((((((.......))).))))))))... ( -34.50) >DroSec_CAF1 108975 88 + 1 UGAUUUGCCAGC------AGA--------G-------CUCCACAUUUGAAUGUGAAUGUGGAACCUGUCGGGCUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ......(((.((------((.--------(-------.(((((((((......))))))))).)))))..))).((..(((((((((.......))).))))))..)). ( -29.90) >DroSim_CAF1 119506 96 + 1 UGAUUUGCCAGC------AGAGGGGCACAG-------CUCCACAUUUGAAUGCGAAUGUGGAACCUGUCGGGCUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ((((..(((.((------((((..(.((((-------(((((((((((....)))))))))).((....))))))))..)))))))))........))))......... ( -35.40) >DroEre_CAF1 108860 95 + 1 UGAUUUGCCAGC------AGAAGGGCGGAGGGGCU--AUCCACAUUUGA------AUGUGGAACCUGUCGGACUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ......((((((------((.....(.((.(((..--.(((((((....------))))))).))).)).).))))))(((((((((.......))).))))))))... ( -31.50) >DroYak_CAF1 109847 97 + 1 UGAUUUGCCAGC------AGAGGGGCACUGCCUCUCCCUUCACAUUUGA------AUGUGGAACCUGUCGGGCUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ......((((((------.(((((((...)))))))..(((((((....------))))))).((....))))))...(((((((((.......))).))))))))... ( -31.10) >DroAna_CAF1 104937 86 + 1 UGAUUUGCUUGC------AGG--GGCA--U-------UUUAACAUUUAA------AUGUGGAACCUGUCAGGCUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ......((((((------(((--.(((--(-------((........))------))))....)))).))))).((..(((((((((.......))).))))))..)). ( -24.90) >consensus UGAUUUGCCAGC______AGAGGGGCA_AG_______CUCCACAUUUGA______AUGUGGAACCUGUCGGGCUGUUGACUCUGCGGUUUUAAUUUGUCAGAGUGCGCU ......((((((..........................(((((((..........))))))).((....))))))...(((((((((.......))).))))))))... (-17.81 = -18.12 + 0.31)


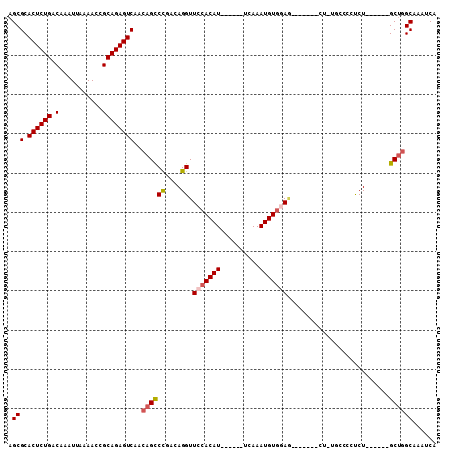
| Location | 9,336,684 – 9,336,781 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9336684 97 - 27905053 AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGCCCAACAGGUUCCACAUUCACAUUCAAAUGUGGAG-------CU-----CCUCUGUUCGAGCUGGCAAAUCA .(((.((((((.(...........))))))))..(((((.(((((((((((((((........)))))))))-------).-----...))))).).))))))...... ( -31.70) >DroSec_CAF1 108975 88 - 1 AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGCCCGACAGGUUCCACAUUCACAUUCAAAUGUGGAG-------C--------UCU------GCUGGCAAAUCA ...(.((((((.(...........))))))))....(((.(.(((((((((((((........)))))))))-------)--------.))------)).)))...... ( -29.30) >DroSim_CAF1 119506 96 - 1 AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGCCCGACAGGUUCCACAUUCGCAUUCAAAUGUGGAG-------CUGUGCCCCUCU------GCUGGCAAAUCA ......................((((((((...(((((((....)).((((((((........)))))))))-------))))....))))------)).))....... ( -28.80) >DroEre_CAF1 108860 95 - 1 AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGUCCGACAGGUUCCACAU------UCAAAUGUGGAU--AGCCCCUCCGCCCUUCU------GCUGGCAAAUCA .(((.((((((.(...........))))))))..((((..((.(((.(((((((------....))))))).--.((......))))))).------))))))...... ( -23.20) >DroYak_CAF1 109847 97 - 1 AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGCCCGACAGGUUCCACAU------UCAAAUGUGAAGGGAGAGGCAGUGCCCCUCU------GCUGGCAAAUCA .(((.((((((.(...........))))))))..((((((....))...(((((------....)))))...((((.(((...))).))))------))))))...... ( -25.90) >DroAna_CAF1 104937 86 - 1 AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGCCUGACAGGUUCCACAU------UUAAAUGUUAAA-------A--UGCC--CCU------GCAAGCAAAUCA ...(.((((((.(...........))))))))....((.((.((((.....(((------((........))-------)--))..--)))------))).))...... ( -15.60) >consensus AGCGCACUCUGACAAAUUAAAACCGCAGAGUCAACAGCCCGACAGGUUCCACAU______UCAAAUGUGGAG_______CU_UGCCCCUCU______GCUGGCAAAUCA .(((.((((((.(...........))))))))..((((((....)).(((((((..........)))))))..........................))))))...... (-17.36 = -17.92 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:15 2006