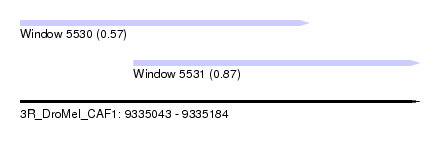
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,335,043 – 9,335,184 |
| Length | 141 |
| Max. P | 0.869873 |

| Location | 9,335,043 – 9,335,145 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -10.92 |
| Energy contribution | -11.42 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9335043 102 + 27905053 AUCAGAGUAGUUUAGUGUGUUACUUUGUUGGUCACCGGCAAACUUUCUCGGCCAGCAG-UUACAUGGGCUUCAAACCUAUGUACAAACUUAUA----UAUAUACACC ..............((((((....(((((((((................)))))))))-.((((((((.......))))))))..........----...)))))). ( -23.59) >DroPse_CAF1 103824 84 + 1 AUCAGAGUAGUUUCGUGUGUUACUUUGUUGGUCGGUGGCAAACUUUGUCGGCCAUCUGUAUAUAUU-------UAUGUA------------CA----UAUAUACAAC ..((((((((.........)))))))).(((((((..(......)..)))))))..(((((((((.-------......------------.)----)))))))).. ( -22.20) >DroSec_CAF1 107341 91 + 1 AUCAGAGUAGUUUAGUGUGUUACUUUGUUGGUCGCUGGCAAACUUUCUCGGCCAGCAG-AU-------CUCCCAACCUAUGUACA----UGCA----UAUAUACACC ......(((.....((((((.((...(((((((((((((...........)))))).)-).-------...)))))....)))))----))).----....)))... ( -23.40) >DroSim_CAF1 116615 98 + 1 AUCAGAGUAGUUUAGUGUGUUACUUUGUUGGUCGCUGGCAAACUUUCUCGGCCAGCAG-AUACAUGGGCUCCCAACCUAUGUACA----UACA----UAUAUACACC ......(((.....(((((....(((((((((((..((....))....))))))))))-)((((((((.......))))))))))----))).----....)))... ( -28.80) >DroEre_CAF1 107252 102 + 1 AUCAGAGUAGUUUAGUGUGUUACUUUGUUGGUCGCCUGCAAACUUUCUCGGCCAGCAG-AUACAUGGGCUCACAACCUAUGUACA----UACAUACUCGUAUAUACC ....(((((.....(((((....(((((((((((..............))))))))))-)((((((((.......))))))))))----))).)))))......... ( -30.84) >DroYak_CAF1 107527 83 + 1 AUCAGAGUAGUUUAGUGUGUUACUUUGUUGGUCGCCGGCAAACUUUCUCGGCCCGCAU-AUAUAU---------------GUAUA----AAUA----UGUAUAUACC ..((((((((.........))))))))..(((.((.(((...........))).))((-((((((---------------((...----.)))----)))))))))) ( -18.20) >consensus AUCAGAGUAGUUUAGUGUGUUACUUUGUUGGUCGCCGGCAAACUUUCUCGGCCAGCAG_AUACAUG__CUCCCAACCUAUGUACA____UACA____UAUAUACACC ...(((((((.........)))))))((((((((..............))))))))................................................... (-10.92 = -11.42 + 0.50)


| Location | 9,335,083 – 9,335,184 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -12.53 |
| Energy contribution | -15.00 |
| Covariance contribution | 2.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9335083 101 + 27905053 AACUUUCUCGGCCAGCAGUUACAUGGGCUUCAAACCUAUGUACAAACUUAUA----UAUAUACACCAGCCCGUAUAUACAGCCUUGGGUAGUUUGUGUGCCAUAU .........(((((.....((((((((.......))))))))((((((....----(((((((........)))))))..(((...))))))))))).))).... ( -24.30) >DroSec_CAF1 107381 90 + 1 AACUUUCUCGGCCAGCAGAU-------CUCCCAACCUAUGUACA----UGCA----UAUAUACACCAGCCCGUAUAUACAGCCUUGGGUAGUUUGUGUGCCAUAU .........(((..((((((-------..(((((....((((.(----(((.----...............)))).))))...)))))..))))))..))).... ( -23.49) >DroSim_CAF1 116655 97 + 1 AACUUUCUCGGCCAGCAGAUACAUGGGCUCCCAACCUAUGUACA----UACA----UAUAUACACCGGCCCGUAUAUACAGCCUUGGGUAGUUUGUGUGCCAUAU .........(((........((((..((((((((..(((((...----.)))----))........(((..((....)).)))))))).)))..))))))).... ( -24.50) >DroEre_CAF1 107292 101 + 1 AACUUUCUCGGCCAGCAGAUACAUGGGCUCACAACCUAUGUACA----UACAUACUCGUAUAUACCAGCCUGUAUAUACAGCCUUGUGUAGUUUGUGCGCCAUAU .........(((..(((((((((((((((.......(((((...----.)))))...((((((((......)))))))))))).))))).))))))..))).... ( -29.00) >DroYak_CAF1 107567 82 + 1 AACUUUCUCGGCCCGCAUAUAUAU---------------GUAUA----AAUA----UGUAUAUACCAGCCCUUAUAUACAGCCUUGGGUAGUUUGUGUGCCAUAU .........(((.(((((((((((---------------((...----.)))----)))))))....((((..............)))).....))).))).... ( -20.04) >consensus AACUUUCUCGGCCAGCAGAUACAUGGGCUCCCAACCUAUGUACA____UACA____UAUAUACACCAGCCCGUAUAUACAGCCUUGGGUAGUUUGUGUGCCAUAU .........(((..(((((((((((((.......)))))))..........................(((((............))))).))))))..))).... (-12.53 = -15.00 + 2.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:13 2006