
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,333,654 – 9,333,776 |
| Length | 122 |
| Max. P | 0.999913 |

| Location | 9,333,654 – 9,333,756 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.48 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9333654 102 + 27905053 ------------AGGGGCACUGUGAGUGGAAGGAGGGGCAGGGGUAGGGAACCCUCACCUUAAGUAAUGAACCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGC ------------....(((...((((((((....((..((((((((((....))).)))))......))..)).((((((.((......)).))))))..))))))))...))) ( -32.40) >DroPse_CAF1 102194 84 + 1 C-----------------------AGCGGGUAG--UGGG--CUGGAGGGG---CUCUCCUUAAGUAAUGAAGCAAGCGCUUGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGC .-----------------------...(.(((.--((.(--((.(((((.---...))))).)))(((((((((.(((((..(......)..)))))))))))))))).))).) ( -33.20) >DroSec_CAF1 105974 82 + 1 -----------------------AAG---------GGGGAGAGGUAGGGGACCCCCACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGC -----------------------...---------.....(((((.(((....))))))))....(((((((((.(((((.((......)).))))))))))))))........ ( -35.30) >DroEre_CAF1 105854 111 + 1 AGCGGGUGGAGCUGGAGCAGCGGGAGCAG---CGGGAGCAGCGGGAGGGUGCCCCAACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGC .((((((...((((...))))(((.((((---(....))..(....)..)))))).))))((...(((((((((.(((((.((......)).))))))))))))))...)).)) ( -44.30) >DroPer_CAF1 106930 91 + 1 C-------------GGG---UGUGAGCGGGUAG--UGGG--GUGGAGGGG---CUCUCCUUAAGUAAUGAAGCAAGCGCUUGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGC .-------------...---.....(((.....--((((--(.((((...---)))))))))...(((((((((.(((((..(......)..)))))))))))))).....))) ( -31.70) >consensus ______________G_G____G_GAGCGG___G__GGGGAGCGGGAGGGG_CCCUCACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGC .............................................((((........))))....(((((((((.(((((.((......)).))))))))))))))........ (-24.04 = -24.24 + 0.20)

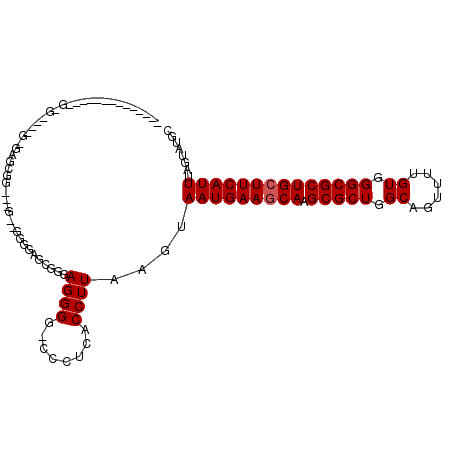

| Location | 9,333,654 – 9,333,756 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.48 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9333654 102 - 27905053 GCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGGUUCAUUACUUAAGGUGAGGGUUCCCUACCCCUGCCCCUCCUUCCACUCACAGUGCCCCU------------ ((((....((((((.((((((...((....))...)))).)).))))))......(((..((((.....))))..)))................))))....------------ ( -27.50) >DroPse_CAF1 102194 84 - 1 GCAUACUAAAUGAAGCAGCGCCCACAAAACUGCAAGCGCUUGCUUCAUUACUUAAGGAGAG---CCCCUCCAG--CCCA--CUACCCGCU-----------------------G ((......(((((((((((((...((....))...)))).)))))))))......((((..---...))))..--....--......)).-----------------------. ( -25.60) >DroSec_CAF1 105974 82 - 1 GCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUGGGGGUCCCCUACCUCUCCCC---------CUU----------------------- ........(((((((((((((...((....))...)))).))))))))).....((((((((...))))))))......---------...----------------------- ( -29.50) >DroEre_CAF1 105854 111 - 1 GCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUUGGGGCACCCUCCCGCUGCUCCCG---CUGCUCCCGCUGCUCCAGCUCCACCCGCU ((......(((((((((((((...((....))...)))).)))))))))......((((((((((.......((.((....)---).))......))))))))))......)). ( -37.82) >DroPer_CAF1 106930 91 - 1 GCAUACUAAAUGAAGCAGCGCCCACAAAACUGCAAGCGCUUGCUUCAUUACUUAAGGAGAG---CCCCUCCAC--CCCA--CUACCCGCUCACA---CCC-------------G ((......(((((((((((((...((....))...)))).)))))))))......((((..---...))))..--....--......)).....---...-------------. ( -25.60) >consensus GCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUGAGGG_CCCCUCCCCCUCCCC__C___CCGCUC_C____C_C______________ ........(((((((((((((...((....))...)))).))))))))).....(((.(.....).)))............................................. (-17.24 = -17.00 + -0.24)



| Location | 9,333,682 – 9,333,776 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9333682 94 + 27905053 GGGGUAGG-GAACCCUCACCUUAAGUAAUGAACCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGCCA--------------GG--CAACAUAGCCCC----AUCU ((((((((-....)))..(((((...((((((...((((((.((......)).))))))..))))))...))....)--------------))--.......)))))----.... ( -31.40) >DroPse_CAF1 102208 106 + 1 -CUGGAGG-GG---CUCUCCUUAAGUAAUGAAGCAAGCGCUUGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGCCACAAACUGAGUUUCAGGUACAAUAUAUCCCU----CUCU -..(((((-((---(....((.....(((((((((.(((((..(......)..)))))))))))))).))...))).....(((.....)))............)))----)).. ( -38.60) >DroSim_CAF1 115261 94 + 1 GGGGUAGG-GGACCCCCACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGCCA--------------GG--CAACAUAGCCCC----AUCU (((((.((-(....))).(((((...(((((((((.(((((.((......)).))))))))))))))...))....)--------------))--.......)))))----.... ( -41.10) >DroEre_CAF1 105891 94 + 1 GCGGGAGG-GUGCCCCAACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGCCG--------------GG--UAACAUAGCCCC----AUCU ((((..((-(...)))..)).((...(((((((((.(((((.((......)).))))))))))))))...)).)).(--------------((--(......)))).----.... ( -36.50) >DroYak_CAF1 106175 95 + 1 GGGGGAGGUGGAUCCCCACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGCCA--------------GG--UAACAUAGCCCC----AUCU (((((((((((....)))))))....(((((((((.(((((.((......)).))))))))))))))...((((...--------------..--...)))).))))----.... ( -43.10) >DroAna_CAF1 102077 89 + 1 ------GG-GG---CUCUCCUUAAGUAAUGAAGCAGGCGCUUGCAGUUUUGCGGGCGCUGCUUCAUUUAGUAUGCCA--------------GG--UUACCCAGCUCCCCUGGUCC ------((-((---((..(((((...((((((((((.(((((((......)))))))))))))))))...))....)--------------))--......))))))........ ( -35.70) >consensus GGGGGAGG_GGACCCCCACCUUAAGUAAUGAAGCAAGCGCUGGCAGUUUUGUGGGCGCUGCUUCAUUUAGUAUGCCA______________GG__CAACAUAGCCCC____AUCU .....(((.((....)).)))((...((((((((.((((((.((......)).))))))))))))))...))........................................... (-23.20 = -23.32 + 0.11)

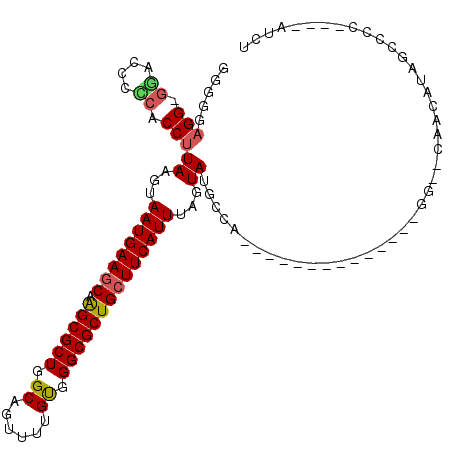
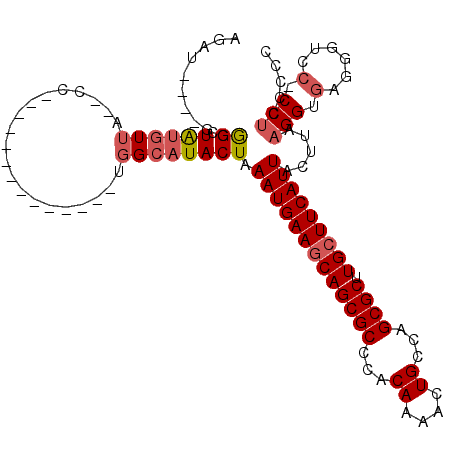
| Location | 9,333,682 – 9,333,776 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -17.28 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9333682 94 - 27905053 AGAU----GGGGCUAUGUUG--CC--------------UGGCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGGUUCAUUACUUAAGGUGAGGGUUC-CCUACCCC ....----((((.((.(..(--((--------------(..(((....((((((.((((((...((....))...)))).)).)))))).......))).))))..-).)))))) ( -27.30) >DroPse_CAF1 102208 106 - 1 AGAG----AGGGAUAUAUUGUACCUGAAACUCAGUUUGUGGCAUACUAAAUGAAGCAGCGCCCACAAAACUGCAAGCGCUUGCUUCAUUACUUAAGGAGAG---CC-CCUCCAG- .(.(----((((.(((.(..((.(((.....)))..))..).)))...(((((((((((((...((....))...)))).)))))))))............---.)-)))))..- ( -29.50) >DroSim_CAF1 115261 94 - 1 AGAU----GGGGCUAUGUUG--CC--------------UGGCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUGGGGGUCC-CCUACCCC .(.(----.((((......)--))--------------).))......(((((((((((((...((....))...)))).)))))))))......(((((((...)-)))))).. ( -34.90) >DroEre_CAF1 105891 94 - 1 AGAU----GGGGCUAUGUUA--CC--------------CGGCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUUGGGGCAC-CCUCCCGC ....----((((...(((..--((--------------((((......(((((((((((((...((....))...)))).))))))))).......))))))))).-..)))).. ( -33.32) >DroYak_CAF1 106175 95 - 1 AGAU----GGGGCUAUGUUA--CC--------------UGGCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUGGGGAUCCACCUCCCCC ....----((((.((((((.--..--------------.))))))...(((((((((((((...((....))...)))).))))))))).....((((((....)))))))))). ( -35.20) >DroAna_CAF1 102077 89 - 1 GGACCAGGGGAGCUGGGUAA--CC--------------UGGCAUACUAAAUGAAGCAGCGCCCGCAAAACUGCAAGCGCCUGCUUCAUUACUUAAGGAGAG---CC-CC------ ......((((.((..((...--))--------------..))......(((((((((((((..(((....)))..))).))))))))))............---))-))------ ( -35.80) >consensus AGAU____GGGGCUAUGUUA__CC______________UGGCAUACUAAAUGAAGCAGCGCCCACAAAACUGCCAGCGCUUGCUUCAUUACUUAAGGUGAGGGUCC_CCUCCCCC ..........((.((((((....................)))))))).(((((((((((((...((....))...)))).))))))))).....(((.(......).)))..... (-17.28 = -18.00 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:07 2006