
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,329,450 – 9,329,566 |
| Length | 116 |
| Max. P | 0.805463 |

| Location | 9,329,450 – 9,329,566 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9329450 116 + 27905053 GGUUUUUUGGGUGGUUUUCGGGUGAUUUUCUGGAGGUGGUACUGGCGGA--CUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAAUGUGCGUUGGUGG ....((((.(((((((.......((((..((((.((.(((...(..(((--(....))))..)..))))).))))..))))))))))).)))).((((((((....)).))))))... ( -37.31) >DroSec_CAF1 100191 102 + 1 GGUUUU------------UGGGUGAUUUUUUGGAGGUGGUA--AGCGGA--CUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAAUGUGCGUUGGUGG (((...------------.((.(((((((((((.((.((..--((.(((--(....))).).))..)))).))))))))))).)))))......((((((((....)).))))))... ( -31.70) >DroSim_CAF1 102485 102 + 1 GGUUUU------------UGGGUGAUUUUCUGGAGGUGGUA--AGCGGA--CUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAAUGUGCGUUGGUGG (((...------------.((.(((((..((((.((.((..--((.(((--(....))).).))..)))).))))..))))).)))))......((((((((....)).))))))... ( -34.40) >DroEre_CAF1 101660 113 + 1 GGUUUU-UGGGUGGCUUUCGGGUGAUUUUCUGGUGGUGGCA--AGCGGC--CUGGUGUCUCGCUCGCCCCAUCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAAUGUGCGUUGGUGG ...(((-(.(((((((.......((((..(((((((.(((.--(((((.--(....).).)))).))))))))))..))))))))))).)))).((((((((....)).))))))... ( -45.41) >DroYak_CAF1 101915 113 + 1 GGUUUU-UGGGUGGCUUUUGGGUGAUUUUCUGGUGGUGGUA--AGCGGA--CUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAAUGUGCGUUGGUGG ...(((-(.(((((((.......((((..(((((((.((..--((.(((--(....))).).))..)))))))))..))))))))))).)))).((((((((....)).))))))... ( -46.81) >DroAna_CAF1 98246 95 + 1 --------GGGUGGC----GAGGGGU-------UGGCGGUA--AGCGGUGAACGGUG--GAGUUGACCUCUCCUGGGAGUUGGCCACCAAAAAUUCAAUGAUUAAUGUGCGUUGGUGG --------.((((((----(((((((-------..((....--.((.(....).)).--..))..)))))))(........)))))))......((((((((....)).))))))... ( -33.90) >consensus GGUUUU__GGGUGGC___UGGGUGAUUUUCUGGAGGUGGUA__AGCGGA__CUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAAUGUGCGUUGGUGG .......................((((((((((.((.(((.....(((...)))(((...)))..))))).))))))))))..(((((((.....((........))....))))))) (-21.38 = -21.47 + 0.09)

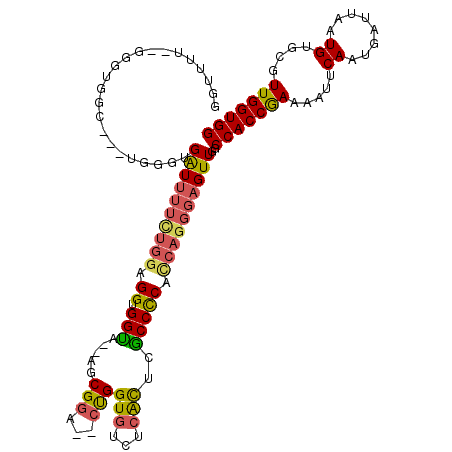

| Location | 9,329,450 – 9,329,566 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -13.06 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9329450 116 - 27905053 CCACCAACGCACAUUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAG--UCCGCCAGUACCACCUCCAGAAAAUCACCCGAAAACCACCCAAAAAACC .........................((((((((((....((....(((((..(..(((.(((....)--)))))..)..)))))...))....))).))))))).............. ( -26.00) >DroSec_CAF1 100191 102 - 1 CCACCAACGCACAUUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAG--UCCGCU--UACCACCUCCAAAAAAUCACCCA------------AAAACC (((((......((........)).......))))).........(((.((((..((((.(((....)--))))))--..)).)).)))............------------...... ( -24.72) >DroSim_CAF1 102485 102 - 1 CCACCAACGCACAUUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAG--UCCGCU--UACCACCUCCAGAAAAUCACCCA------------AAAACC (((((......((........)).......)))))........((((.((((..((((.(((....)--))))))--..)).)).))))...........------------...... ( -27.82) >DroEre_CAF1 101660 113 - 1 CCACCAACGCACAUUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGAUGGGGCGAGCGAGACACCAG--GCCGCU--UGCCACCACCAGAAAAUCACCCGAAAGCCACCCA-AAAACC .........................((((.((((((.......(((.(((((((((((.(.......--.)))))--)))).))).)))....((....))..)))))).)-)))... ( -33.70) >DroYak_CAF1 101915 113 - 1 CCACCAACGCACAUUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAG--UCCGCU--UACCACCACCAGAAAAUCACCCAAAAGCCACCCA-AAAACC .........................((((.((((((.......(((((((((..((((.(((....)--))))))--..)).)))))))..............)))))).)-)))... ( -35.00) >DroAna_CAF1 98246 95 - 1 CCACCAACGCACAUUAAUCAUUGAAUUUUUGGUGGCCAACUCCCAGGAGAGGUCAACUC--CACCGUUCACCGCU--UACCGCCA-------ACCCCUC----GCCACCC-------- ..............................((((((...(((....))).(((.(((..--....))).)))...--........-------.......----)))))).-------- ( -19.80) >consensus CCACCAACGCACAUUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAG__UCCGCU__UACCACCACCAGAAAAUCACCCA___GCCACCC__AAAACC (((((......((........)).......)))))..........(((((((((.................))))....))))).................................. (-13.06 = -13.45 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:03 2006