
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,325,469 – 9,325,569 |
| Length | 100 |
| Max. P | 0.828736 |

| Location | 9,325,469 – 9,325,569 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.63 |
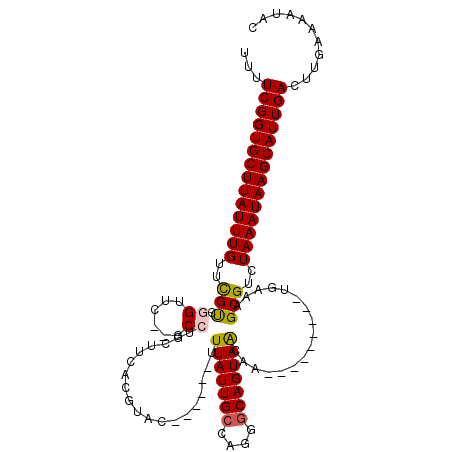
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -23.47 |
| Energy contribution | -23.55 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828736 |
| Prediction | RNA |
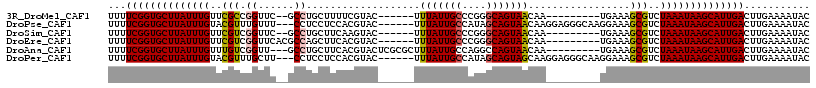
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9325469 100 - 27905053 UUUUCGGUGCUUAUUUGUUCGCCGGUUC--GCCUGCUUUUCGUAC------UUUAUUGCCCGGGCAGUAACAA---------UGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((..(((.((...--.))....((((((..------.(((((((....)))))))..)---------))))))))..))))))))))))))........... ( -33.20) >DroPse_CAF1 93259 108 - 1 UUUUCGGUGCUUAUUUGUACGUUUGUUU---CCUCCUCCACGUAC------UUUAUUGCCAUAGCAGUAACAAGGAGGGCAAGGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((.((((...(((---((((((((..((..------..((((((....))))))))..)))))...)))))))))).))))))))))))))........... ( -38.00) >DroSim_CAF1 98559 100 - 1 UUUUCGGUGCUUAUUUGUUCGUCGGUUC--GCCUGCUUCAAGUAC------UUUAUUGCCCGGGCAGUAACAA---------UGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((..(...((...--.)).((((((.....------.(((((((....)))))))...---------)).)))))..))))))))))))))........... ( -28.10) >DroEre_CAF1 97826 102 - 1 UUUUCGGUGCUUAUUUGUUCGUCGGUUCACGCCAGCUUCACGUAC------UUUAUUGCCCGGGCAGUAACAA---------UGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((..(...(((....))).((((..(((..------.(((((((....)))))))..)---------)).)))))..))))))))))))))........... ( -30.00) >DroAna_CAF1 94337 105 - 1 UUUUCGGUGCUUAUUUGUUUGUCGGUU---GCCUGCUUCACGUACUCGCGCUUUAUUGCCAGGCCAGUAACAA---------UGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((..(((..(((---((..((((..((....)).((......)).))))..)))))..---------.....)))..))))))))))))))........... ( -24.70) >DroPer_CAF1 97010 108 - 1 UUUUCGGUGCUUAUUUGUACGUUUGCUU---CCUCCUCCACGUAC------UUUAUUGCCAUAGCAGUAGCAAGGAGGGCAAGGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((.((((((.(((---..((((((..((..------..((((((....))))))))..))))))..))).)))))).))))))))))))))........... ( -39.20) >consensus UUUUCGGUGCUUAUUUGUUCGUCGGUUC__GCCUGCUUCACGUAC______UUUAUUGCCAGGGCAGUAACAA_________UGAAAGCGUCUAAAUAAGCAUUGACUUGAAAAUAC ...((((((((((((((..(((.((......))...................(((((((....))))))).................)))..))))))))))))))........... (-23.47 = -23.55 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:58 2006