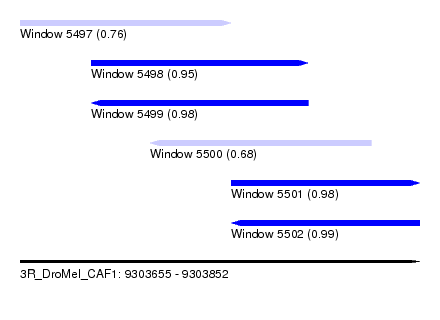
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,303,655 – 9,303,852 |
| Length | 197 |
| Max. P | 0.991277 |

| Location | 9,303,655 – 9,303,759 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.81 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9303655 104 + 27905053 CAACUCAAGGAGCUGCCCUCGUUGGGCAGUAAAACUGGCGCCCAACAG-----AGGAUGGCAUUGUUGGGCGUCACCAUGACCAUCAAUAUGAACGGAAUGCGAGGUAG ...(((..((.(((((((.....))))))).....(((((((((((((-----.(.....).)))))))))))))))....((.((.....))..)).....))).... ( -41.20) >DroPse_CAF1 72467 105 + 1 UAACUUUGCAAGCGGCCCUCGG-GGGCCGUAAAACUGCCGGCAAACAGCGUAGGGGCAGGCAUUUCUGGGUGGCGUCAUGACCAUCAAUAUGAACGGAAUGCCAGG--- ...((((((..(((((((....-)))))))......((.(.....).))))))))...((((((((..(((((........))))).........))))))))...--- ( -37.10) >DroSim_CAF1 76157 104 + 1 CAACUCAAGGAGCUGCCCUCGUUGGGCAGUAAAACUGGCGCCCAACAG-----AGGAUGGCAUUGUUGGGCGGCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUAG ...(((..((.(((((((.....))))))).....((.((((((((((-----.(.....).)))))))))).))))....((.((.....))..)).....))).... ( -40.90) >DroEre_CAF1 75664 104 + 1 CAACUCAAGGAGCUGCCCUCGCCGGGCAGUAAAACUGGCGCCCAACAG-----AGGAUGGCAUUGUUGGGCGUCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUUG ...(((..((.(((((((.....))))))).....(((((((((((((-----.(.....).)))))))))))))))....((.((.....))..)).....))).... ( -42.20) >DroYak_CAF1 75585 104 + 1 CAACUGAAGGAGCUGCCCUCGCUGGGCAGUACAACUGGCGCCCAACAG-----AGGACGGCAUUGUUGGGCGUCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUAG ........((.(((((((.....))))))).....(((((((((((((-----.(.....).)))))))))))))))...(((.((.....))((.....))..))).. ( -40.40) >DroPer_CAF1 75663 105 + 1 UAACUUUGCAAACGGCCCUCGG-GGUCCGUAAAACUGCCGGCAAACAGCGUAGGGGCAGGCAUUUCUGGGUGGCGUCAUGACCAUCAAUAUGAACGGAAUGCCAGG--- ...((((((..(((((((...)-)).))))......((.(.....).))))))))...((((((((..(((((........))))).........))))))))...--- ( -31.10) >consensus CAACUCAAGGAGCUGCCCUCGCUGGGCAGUAAAACUGGCGCCCAACAG_____AGGAUGGCAUUGUUGGGCGGCACCAUGACCAUCAAUAUGAACGGAAUGCGAGGUAG ...........(((((((.....))))))).....(((((((((((((..............))))))))))))).(((..((.((.....))..)).)))........ (-27.50 = -27.81 + 0.31)


| Location | 9,303,690 – 9,303,797 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9303690 107 + 27905053 UGGCGCCCAA------CAGAGGAUGGCAUUGUUGGGCGUCACCAUGACCAUCAAUAUGAACGGAAUGCGAGGUAGGAGACCGAUGGUGGAUGGUAGAUGGAGAAUGAAGGGCA ((((((((((------(((.(.....).)))))))))))))((((.((((((..(((.(.(((..(.(......).)..))).).)))))))))..))))............. ( -39.00) >DroSec_CAF1 74846 107 + 1 UGGCGCCCAA------CAGAGGAUGGCAUUGUUGGGCGGCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUAGGAGACCGAUGGUGGAUGGUAGGAGGAGAAUGAAGGGCA .(.(((((((------(((.(.....).)))))))))).)(((((.((((((.......((.....))..(((.....))))))))).).))))................... ( -36.60) >DroSim_CAF1 76192 107 + 1 UGGCGCCCAA------CAGAGGAUGGCAUUGUUGGGCGGCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUAGGAGACCGAUGGUGGAUGGUAGGUGGAGAAUGAAGGGCA ((.(((((((------(((.(.....).)))))))))).))((((.((((((.((.....(((..(.(......).)..)))...)).))))))..))))............. ( -37.50) >DroEre_CAF1 75699 93 + 1 UGGCGCCCAA------CAGAGGAUGGCAUUGUUGGGCGUCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUUGGAGACCG-------GUGGUGGAU-------GGAGGGCA ((((((((((------(((.(.....).)))))))))))))((((.((((((.......((.....))..((((...)))))-------))))).).)-------))...... ( -37.30) >DroYak_CAF1 75620 100 + 1 UGGCGCCCAA------CAGAGGACGGCAUUGUUGGGCGUCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUAGGAGACCGAUGGUUGGUGGUAGAU-------GGAGGGCA ((((((((((------(((.(.....).)))))))))))))((((.(((((((((.....(((..(.(......).)..)))...)))))))))..))-------))...... ( -42.60) >DroAna_CAF1 73689 94 + 1 UCCCUACCGAAUAGCUCAGAGGCCGGCAUUGUUGGGAGGCGCCAGGACCAUCAAUAUGACCAGAAUCCAAGGUGGGGGACG-AAGGC-GAUGGUUG----------------- ((((((((.....(((....))).(((((.(((((..(((.....).)).)))))))).)).........))))))))...-.....-........----------------- ( -23.70) >consensus UGGCGCCCAA______CAGAGGAUGGCAUUGUUGGGCGGCACCAUGACCAUCAAUAUGAGCGGAAUGCGAGGUAGGAGACCGAUGGUGGAUGGUAGAU_______GAAGGGCA ((((((((((......((.....))......)))))))))).....((((((........(((..(.(......).)..)))......))))))................... (-21.04 = -21.46 + 0.42)



| Location | 9,303,690 – 9,303,797 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -16.91 |
| Energy contribution | -18.42 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9303690 107 - 27905053 UGCCCUUCAUUCUCCAUCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGUUCAUAUUGAUGGUCAUGGUGACGCCCAACAAUGCCAUCCUCUG------UUGGGCGCCA ..................((((((..(((((((((.....)))..((.....)).......)))))).)))))).(((((((((..(.....)..))------)))))))... ( -28.70) >DroSec_CAF1 74846 107 - 1 UGCCCUUCAUUCUCCUCCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGCCGCCCAACAAUGCCAUCCUCUG------UUGGGCGCCA ..................((((((..(((((((((.....)))..((.....)).......)))))).)))))).(((((((((..(.....)..))------)))))))... ( -30.70) >DroSim_CAF1 76192 107 - 1 UGCCCUUCAUUCUCCACCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGCCGCCCAACAAUGCCAUCCUCUG------UUGGGCGCCA ..................((((((..(((((((((.....)))..((.....)).......)))))).)))))).(((((((((..(.....)..))------)))))))... ( -30.70) >DroEre_CAF1 75699 93 - 1 UGCCCUCC-------AUCCACCAC-------CGGUCUCCAACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGACGCCCAACAAUGCCAUCCUCUG------UUGGGCGCCA .(((..((-------(((......-------.(((.....)))..((.....)).......)))))....)))(.(((((((((..(.....)..))------))))))).). ( -26.50) >DroYak_CAF1 75620 100 - 1 UGCCCUCC-------AUCUACCACCAACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGACGCCCAACAAUGCCGUCCUCUG------UUGGGCGCCA ........-------......((((((((((((((.....)))..((.....)).......))))))..))))).(((((((((..(.....)..))------)))))))... ( -32.40) >DroAna_CAF1 73689 94 - 1 -----------------CAACCAUC-GCCUU-CGUCCCCCACCUUGGAUUCUGGUCAUAUUGAUGGUCCUGGCGCCUCCCAACAAUGCCGGCCUCUGAGCUAUUCGGUAGGGA -----------------..((((((-(((..-.((((........))))...)))......)))))).((((((...........))))))((((((((...))))).))).. ( -23.80) >consensus UGCCCUUC_______AUCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGACGCCCAACAAUGCCAUCCUCUG______UUGGGCGCCA ..................(((((...(((((((((.....)))..((.....)).......))))))..))))).(((((((......((.....))......)))))))... (-16.91 = -18.42 + 1.50)



| Location | 9,303,719 – 9,303,828 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -18.30 |
| Energy contribution | -20.33 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9303719 109 - 27905053 GCACGGUCUG-UCAGCUGGCCACAUGGCCAUCUGCCCUUCAUUCUCCAUCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGUUCAUAUUGAUGGUCAUGGUGACGC .....(((..-.(((.(((((....))))).))).................(((((..(((((((((.....)))..((.....)).......)))))).)))))))).. ( -27.80) >DroSec_CAF1 74875 109 - 1 GCAGGGUCUG-UCAGCUGGCCAGAUGGCCAUCUGCCCUUCAUUCUCCUCCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGCCGC ((((((....-.(((.(((((....))))).)))))))............((((((..(((((((((.....)))..((.....)).......)))))).))))))..)) ( -33.00) >DroSim_CAF1 76221 109 - 1 GCAGGGUCUG-UCAGCUGGCCAGAUGGCCAUCUGCCCUUCAUUCUCCACCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGCCGC ((((((....-.(((.(((((....))))).)))))))............((((((..(((((((((.....)))..((.....)).......)))))).))))))..)) ( -33.00) >DroEre_CAF1 75728 95 - 1 GCAGUGUCUG-UCAGCUGGCCAGAUGGCCAACUGCCCUCC-------AUCCACCAC-------CGGUCUCCAACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGACGC ...(((((..-.(((.(((((....))))).))).((.((-------(((......-------.(((.....)))..((.....)).......)))))....)).))))) ( -27.00) >DroYak_CAF1 75649 102 - 1 GCAGUGUCUG-UCAGCUGGCCAGAUGGCCAACUGCCCUCC-------AUCUACCACCAACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGACGC ...(((((..-.(((.(((((....))))).)))......-------.......(((((((((((((.....)))..((.....)).......))))))..))))))))) ( -31.90) >DroAna_CAF1 73724 91 - 1 GCUGAGUCUGGCCAUCUGGCCAUUUGGUCAUC-----------------CAACCAUC-GCCUU-CGUCCCCCACCUUGGAUUCUGGUCAUAUUGAUGGUCCUGGCGCCUC ........(((((....)))))...(((...(-----------------((((((((-(((..-.((((........))))...)))......))))))..))).))).. ( -26.00) >consensus GCAGGGUCUG_UCAGCUGGCCAGAUGGCCAUCUGCCCUUC_______AUCUACCAUCCACCAUCGGUCUCCUACCUCGCAUUCCGCUCAUAUUGAUGGUCAUGGUGACGC ............(((.(((((....))))).)))................(((((...(((((((((.....)))..((.....)).......))))))..))))).... (-18.30 = -20.33 + 2.03)

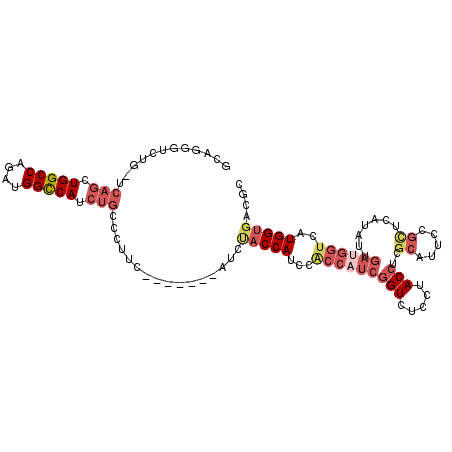
| Location | 9,303,759 – 9,303,852 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -21.16 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9303759 93 + 27905053 GAGACCGAUGGUGGAUGGUAGAUGGAGAAUGAAGGGCAGAUGGCCAUGUGGCCAGCUGACAGACCGUGCUCCUCCUUCCUUCCAAACGGCAGC ..(.(((....((((.((.((..((((.(((....((((.(((((....))))).))).)....))).))))..)).)).))))..))))... ( -34.60) >DroSec_CAF1 74915 91 + 1 GAGACCGAUGGUGGAUGGUAGGAGGAGAAUGAAGGGCAGAUGGCCAUCUGGCCAGCUGACAGACCCUGCUCCUCCUUCCAC--CAACGGCAGC ..(.(((.(((((((....((((((((.....(((((((.(((((....))))).))).....)))).)))))))))))))--)).))))... ( -48.20) >DroSim_CAF1 76261 91 + 1 GAGACCGAUGGUGGAUGGUAGGUGGAGAAUGAAGGGCAGAUGGCCAUCUGGCCAGCUGACAGACCCUGCUCCUCCUUCCAC--CAACGGCAGC ..(.(((.(((((((....(((.((((.....(((((((.(((((....))))).))).....)))).)))).))))))))--)).))))... ( -44.00) >DroEre_CAF1 75768 77 + 1 GAGACCG-------GUGGUGGAU-------GGAGGGCAGUUGGCCAUCUGGCCAGCUGACAGACACUGCUCCUCCUCCCUC--CAACGGCUGC .((.(((-------.(((.((..-------(((((((((((((((....))))))))).(((...))).))))))..)).)--)).))))).. ( -40.00) >DroYak_CAF1 75689 84 + 1 GAGACCGAUGGUUGGUGGUAGAU-------GGAGGGCAGUUGGCCAUCUGGCCAGCUGACAGACACUGCUCCUCCUCCCCA--UAACGUCUGC ......((((..(((.((..((.-------((((..(((((((((....))))))))).(((...)))))))))..)))))--...))))... ( -35.60) >consensus GAGACCGAUGGUGGAUGGUAGAUGGAGAAUGAAGGGCAGAUGGCCAUCUGGCCAGCUGACAGACCCUGCUCCUCCUUCCAC__CAACGGCAGC ...((((........))))...........(((((((((.(((((....))))).))).(((...)))....))))))............... (-21.16 = -20.88 + -0.28)

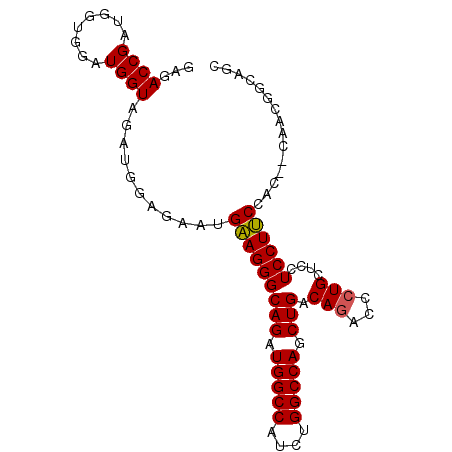

| Location | 9,303,759 – 9,303,852 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9303759 93 - 27905053 GCUGCCGUUUGGAAGGAAGGAGGAGCACGGUCUGUCAGCUGGCCACAUGGCCAUCUGCCCUUCAUUCUCCAUCUACCAUCCACCAUCGGUCUC ...((((..((((.((.((..((((...((...(.(((.(((((....))))).))))...))...))))..)).)).))))....))))... ( -32.80) >DroSec_CAF1 74915 91 - 1 GCUGCCGUUG--GUGGAAGGAGGAGCAGGGUCUGUCAGCUGGCCAGAUGGCCAUCUGCCCUUCAUUCUCCUCCUACCAUCCACCAUCGGUCUC ...((((.((--(((((((((((((.((((.....(((.(((((....))))).))))))).....))))))))....))))))).))))... ( -46.90) >DroSim_CAF1 76261 91 - 1 GCUGCCGUUG--GUGGAAGGAGGAGCAGGGUCUGUCAGCUGGCCAGAUGGCCAUCUGCCCUUCAUUCUCCACCUACCAUCCACCAUCGGUCUC ...((((.((--((((((((.((((.((((.....(((.(((((....))))).))))))).....)))).)))....))))))).))))... ( -42.00) >DroEre_CAF1 75768 77 - 1 GCAGCCGUUG--GAGGGAGGAGGAGCAGUGUCUGUCAGCUGGCCAGAUGGCCAACUGCCCUCC-------AUCCACCAC-------CGGUCUC ...((((.((--(.((..(((((.(((((((......))(((((....)))))))))))))))-------..)).))).-------))))... ( -39.30) >DroYak_CAF1 75689 84 - 1 GCAGACGUUA--UGGGGAGGAGGAGCAGUGUCUGUCAGCUGGCCAGAUGGCCAACUGCCCUCC-------AUCUACCACCAACCAUCGGUCUC ..(((((((.--(((((((((((.(((((((......))(((((....)))))))))))))))-------.))).)))..))).....)))). ( -31.90) >consensus GCUGCCGUUG__GAGGAAGGAGGAGCAGGGUCUGUCAGCUGGCCAGAUGGCCAUCUGCCCUUCAUUCUCCAUCUACCAUCCACCAUCGGUCUC ..................(((((.(((((((......))(((((....)))))))))))))))...........(((..........)))... (-22.62 = -22.10 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:45 2006