
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,302,693 – 9,302,797 |
| Length | 104 |
| Max. P | 0.726816 |

| Location | 9,302,693 – 9,302,797 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
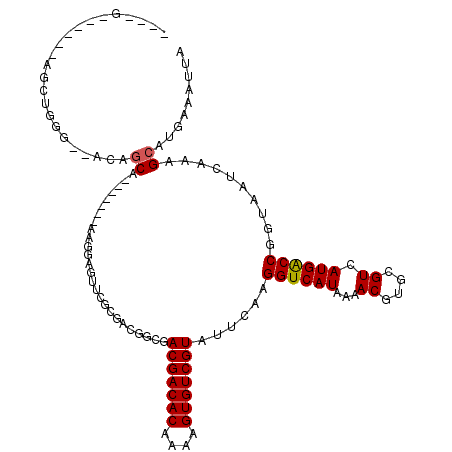
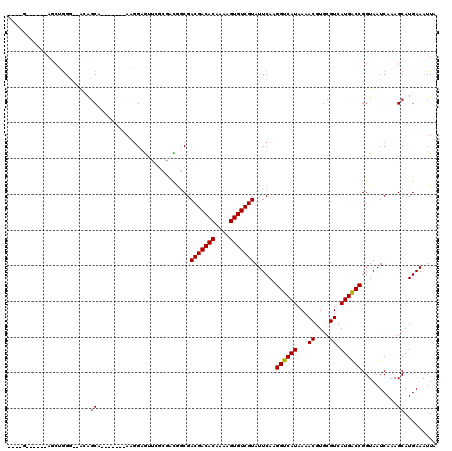
>3R_DroMel_CAF1 9302693 104 + 27905053 ----G------AGCUGGG--GGAGCA----AUGAAGGAGUCCUGGACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUA ----.------.(((..(--(..((.----.((((((.(((...))).)).(((((((....))))))).)))).((((((...((....)).)))))).))..))..)))......... ( -29.60) >DroPse_CAF1 71481 100 + 1 ----G------AGCAGG---ACAGCG-------AUGGCAUUGGCGUUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUA ----.------.(((.(---...((.-------...))..((((.((((..(((((((....)))))))..)))).)))).....).))).(((((....(....)....)))))..... ( -29.60) >DroEre_CAF1 74662 101 + 1 ----U------AUCUGGG--GGAGCA-------AAGGAGUCCUGGACAUCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUA ----.------.((..((--(...(.-------...)..)))..)).(((.(((((((....)))))))......((((((...((....)).))))))))).................. ( -23.30) >DroWil_CAF1 74325 103 + 1 ----U------AAAUGGGCAAUGGCA-------AAGGCAAUGGCACUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUA ----.------..((.((..(((((.-------...((((((((..(((..(((((((....)))))))..)))..)))))......))))))))..)).)).................. ( -26.00) >DroAna_CAF1 72772 112 + 1 ACCAG------CGCCAGG--ACAUCAGAUCAGGAGUGAGCUCGGUGCGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUA (((.(------((((..(--((....).))..(((....))))))))((..(((((((....)))))))..))..((((((...((....)).))))))))).................. ( -31.80) >DroPer_CAF1 74572 106 + 1 ----GAGCAGGAGCAGG---ACAGCG-------AUGGCGAUGGCGUUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUA ----..((.((..((.(---((.((.-------(((...(((((.((((..(((((((....)))))))..)))).)))))....)))))))).)).)).)).................. ( -31.50) >consensus ____G______AGCUGGG__ACAGCA_______AAGGAGUUCGCGACGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUA .......................((..........................(((((((....)))))))......((((((...((....)).))))))..........))......... (-18.95 = -18.90 + -0.05)

| Location | 9,302,693 – 9,302,797 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -17.63 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
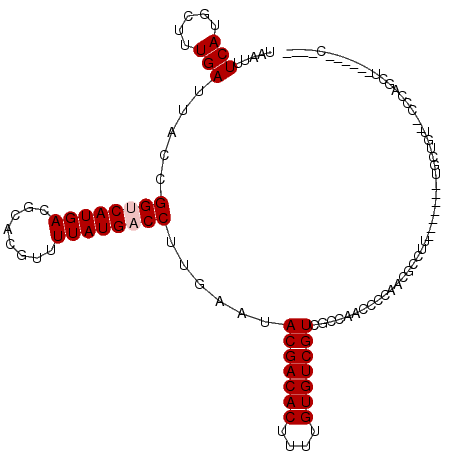
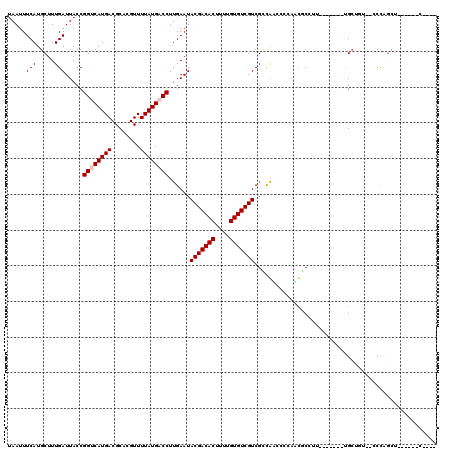
>3R_DroMel_CAF1 9302693 104 - 27905053 UAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGGUGUCCAGGACUCCUUCAU----UGCUCC--CCCAGCU------C---- .........((..(((.......((..(((((.((.((((......)))).(((((((....))))))))))))))..))......))).----.))...--.......------.---- ( -23.62) >DroPse_CAF1 71481 100 - 1 UAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAACGCCAAUGCCAU-------CGCUGU---CCUGCU------C---- .........((.(((......((((((((........))))))))(((...(((((((....)))))))...)))...))).))...-------.((...---...)).------.---- ( -24.20) >DroEre_CAF1 74662 101 - 1 UAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGAUGUCCAGGACUCCUU-------UGCUCC--CCCAGAU------A---- .....(((((((.........)).))))).(((.(((((..(((.((((...((((((....)))))))))).))).))))).....-------)))...--.......------.---- ( -19.80) >DroWil_CAF1 74325 103 - 1 UAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAGUGCCAUUGCCUU-------UGCCAUUGCCCAUUU------A---- .........((..........((((((((........))))))))......(((((((....))))))).))(((((.((.......-------)).))))).......------.---- ( -24.10) >DroAna_CAF1 72772 112 - 1 UAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCGCACCGAGCUCACUCCUGAUCUGAUGU--CCUGGCG------CUGGU .....(((.....)))..(((((((((((........))))))))......(((((((....)))))))(((((((.(.((.(((....))).))).)))--...))))------..))) ( -29.20) >DroPer_CAF1 74572 106 - 1 UAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAACGCCAUCGCCAU-------CGCUGU---CCUGCUCCUGCUC---- .........((.(((......((((((((........))))))))......(((((((....)))))))...))).))....((...-------.((...---...))....))..---- ( -24.70) >consensus UAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAACCCCAACGCCUU_______UGCUGU__CCCAGCU______C____ .....(((.....))).....((((((((........))))))))......(((((((....)))))))................................................... (-17.63 = -17.97 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:39 2006