
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,257,876 – 9,258,022 |
| Length | 146 |
| Max. P | 0.747647 |

| Location | 9,257,876 – 9,257,982 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -34.98 |
| Energy contribution | -35.23 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
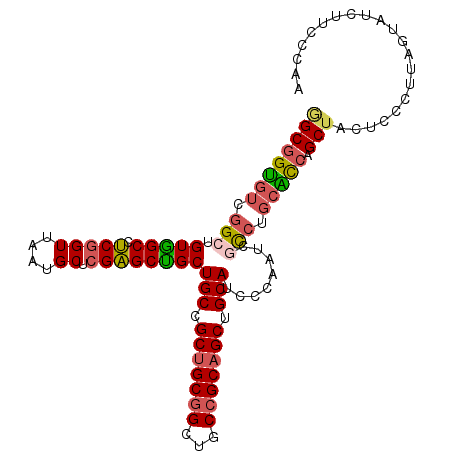
>3R_DroMel_CAF1 9257876 106 - 27905053 GGCGGUGUCGGCUGUGGCCUCGGUUAAUGCUCGAGCUGCUGCCGCUGCGGCUGCCGCAGCCGCAUCCCAAUCGCCUGCACCGGCUACUCCCUUAGUAUCUUCCCAA ((.((((.((((((((((((((((....)).))))..(((((....))))).))))))))))))))))....(((......)))((((.....))))......... ( -45.10) >DroVir_CAF1 30439 99 - 1 GGCAGCCUCUGCCGUUGCCACGGUCAAUGCACGUGCAGCUGCCGCUGCAGCUGCAGCCGCCGCAGCUCAAGUGUC---GUCAGCGGCAUCUGUUGUUCCUUC---- (((((...)))))(((((..((((.........(((((((((....)))))))))))))..))))).((..((((---(....)))))..))..........---- ( -41.70) >DroSec_CAF1 28767 106 - 1 GGCGGUGUCGGCGGUGGCCUCCGUUAAUGCUCGAGCUGCUGCCGCUGCGGCUGCCGCAGCUGCAUCUCAAUCGCCUGCACCGGCUACUCCCUUAGUAUCUUCCCAA ((((((((.(((((..((.((.((....))..))))..)(((.(((((((...))))))).))).......)))).))))))((((......))))......)).. ( -43.00) >DroSim_CAF1 28842 106 - 1 GGCGGUGUCGGCGGUAGCCUCGGUUAAUGCUCGAGCUGCUGCCGCUGCGGCUGCCGCAGCUGCAUCUCAAUCGCCUGCACCGGCUACUCCCUUAGUAUCUUCCCAA ((((((((.(((((((((.(((((....)).))))))))(((.(((((((...))))))).))).......)))).))))))((((......))))......)).. ( -48.00) >DroEre_CAF1 29293 106 - 1 AGCGGUGUCGGCUGUGGCCUCGGUUAAUGCUCGAGCUGCUGCCGCUGCGGCUGCCGCAGCUGCAUCCCAAUCACCUGCGCCAGCUACUCCCUUAAUAUCUUCCCAA ((((((((.((..(..((.(((((....)).)))))..)(((.(((((((...))))))).))).........)).))))).)))..................... ( -38.10) >DroYak_CAF1 28730 106 - 1 GGCGGUGUCGGCUGUGGCCUCGGUCAAUGCUCGAGCUGCUGCCGCUGCGGCUGCCGCAGCUGCAACCCAAUCACCUGCACCAGCUACUCCCUUAGUAUCUUCCCAA ((((((((.((..(..((.(((((....)).)))))..)(((.(((((((...))))))).))).........)).))))).)))..................... ( -37.90) >consensus GGCGGUGUCGGCUGUGGCCUCGGUUAAUGCUCGAGCUGCUGCCGCUGCGGCUGCCGCAGCUGCAUCCCAAUCGCCUGCACCAGCUACUCCCUUAGUAUCUUCCCAA ((((((((.(((.(((((.(((((....)).))))))))(((.(((((((...))))))).)))........))).))))).)))..................... (-34.98 = -35.23 + 0.26)

| Location | 9,257,914 – 9,258,022 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.01 |
| Mean single sequence MFE | -48.57 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.67 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9257914 108 - 27905053 CGCACCUGUUGCGCGAACGUCAAAAGGCUUCCCAGGAGGCGGCGGUGUCGGCUGUGGCCUCGGUUAAUGCUCGAGCUGCUGC---CGCUGCGGCUGCCGCAGCCGCAUCC---C .((.(((.(((.(.(((.(((....)))))))))).)))..))((((.((((((((((((((((....)).))))..(((((---....))))).)))))))))))))).---. ( -50.10) >DroVir_CAF1 30470 108 - 1 CGCAUUUAUUGCGCGAACGCCAGAAGGCUUCGCAGGAAUCGGCAGCCUCUGCCGUUGCCACGGUCAAUGCACGUGCAGCUGC---CGCUGCAGCUGCAGCCGCCGCAGCU---C .(((((.((((.(((((.(((....)))))))).((...((((((...))))))...)).)))).)))))...((((((...---.))))))(((((.......))))).---. ( -49.40) >DroPse_CAF1 27305 111 - 1 CCCAUCUUCUGCGAGAGCGGCAAAAAGCCUCCCAAGAGUCGGCCGCUUCAGCCGUGGCCUCGGUCAAUGCUCGGGCUGCUGCAGCCGCCGCCGCUGCUGCAGCUGCCUCA---C ..............(((((((.....)))......((.(.((((((.......))))))..).))...))))((((.((((((((.((....)).)))))))).))))..---. ( -47.60) >DroMoj_CAF1 29854 99 - 1 CGCACUUGUUGCGCGAGCGUCAAAAG---------GAGUCGGCCGCAGCUGCCGUAGCUACGGUCAAUGCACGAGCAGCAGC---CGCUGCAGCUGCAGCAGCCGCAGGU---C .....(((.(((....))).)))...---------.....((((((.(((((.((((((.((((....)).)).(((((...---.))))))))))).))))).)).)))---) ( -42.20) >DroAna_CAF1 28474 111 - 1 CGCACCUGUUGCGGGAACGCCAAAAGGCCUCCCAGGAAUCGGCUGCCUCCGCUGUGGCUUCAGUUAAUGCCAGAGCUGCCGC---UGCUGCGGCGGCAGCUGCUGCUUCCUCAC .(((.....)))((((..(((....))).))))(((((.((((((((...(((.((((..........)))).))).(((((---....)))))))))))))....)))))... ( -54.30) >DroPer_CAF1 29866 111 - 1 CCCAUCUUCUGCGAGAGCGGCAAAAGGCCUCCCAAGAGUCGGCCGCUUCAGCCGUGGCCUCGGUCAAUGCUCGGGCUGCUGCAGCCGCCGCCGCUGCUGCAGCUGCCUCA---C ...........((((((((((....(((((....)).))).)))))))..((..((((....))))..)))))(((.((((((((.((....)).)))))))).)))...---. ( -47.80) >consensus CGCACCUGUUGCGCGAACGCCAAAAGGCCUCCCAGGAGUCGGCCGCCUCAGCCGUGGCCUCGGUCAAUGCUCGAGCUGCUGC___CGCUGCAGCUGCAGCAGCCGCAUCA___C ....(((.(((((....)).))).)))............((((.((..((((.(((((....((....((....))....))....))))).))))..)).))))......... (-24.80 = -24.67 + -0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:23 2006