
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,202,764 – 9,202,982 |
| Length | 218 |
| Max. P | 0.987395 |

| Location | 9,202,764 – 9,202,865 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -22.15 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202764 101 + 27905053 GCGUAAGGAUCCCCAGGAAAACCAUGAACAUUGUGGACACCACGGCCAUCAUUGGCAUCAUGGGCAUCAUGGAUUUCAUGGACACCAUGGUCACCAUGGAC ((....((....)).......((((((....(((((...)))))((((....)))).)))))))).((((((...((((((...))))))...)))))).. ( -35.00) >DroSec_CAF1 38383 101 + 1 GCGUAAGGAUCCCCAGGAAAACCAUGAGCAUCAUGGACACCAUGGACACCAUUGGCAUCAUGGACAUCAUGGAUUUCAUGGUUACCAUGGCCACCAUAGCC ((.((.((....(((((....(((((.....)))))...)).))).......((((..(((((..((((((.....))))))..))))))))))).)))). ( -29.70) >DroSim_CAF1 37203 101 + 1 GCGUAAGGAUCCCCAGGAAAACCAUGAGCAUCAUGGACACCACGGACACCAUUGGCAUCAUGGACAUCAUGGAUUUCAUGGUUACCAUGGCCACCACGGAC .(((..((.(((...((....(((((.....)))))...))..)))..))..((((..(((((..((((((.....))))))..)))))))))..)))... ( -29.30) >DroYak_CAF1 37296 92 + 1 CCGUAAGGAACACCAUGAAGACCACGGACACCAUGGACAUCACGAACACCCUUGGCAUCAUGGACACCAUGGACAUCAUGGACACC---------CCUGGC ((....))....((((((.(.(((.((...((((((...(((.(......).)))..))))))...)).))).).)))))).....---------...... ( -24.30) >consensus GCGUAAGGAUCCCCAGGAAAACCAUGAACAUCAUGGACACCACGGACACCAUUGGCAUCAUGGACAUCAUGGAUUUCAUGGACACCAUGGCCACCACGGAC ............(((.((((.((((((...((((((...(((..........)))..))))))...)))))).)))).)))...((((((...)))))).. (-22.15 = -22.52 + 0.38)



| Location | 9,202,764 – 9,202,865 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -25.29 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202764 101 - 27905053 GUCCAUGGUGACCAUGGUGUCCAUGAAAUCCAUGAUGCCCAUGAUGCCAAUGAUGGCCGUGGUGUCCACAAUGUUCAUGGUUUUCCUGGGGAUCCUUACGC ..((((((...))))))(.((((.((((.((((((.(((((((..((((....))))))))).)).........)))))).)))).)))).)......... ( -32.30) >DroSec_CAF1 38383 101 - 1 GGCUAUGGUGGCCAUGGUAACCAUGAAAUCCAUGAUGUCCAUGAUGCCAAUGGUGUCCAUGGUGUCCAUGAUGCUCAUGGUUUUCCUGGGGAUCCUUACGC ((((.....))))..(((..(((.((((.((((((.((.((((((((((.(((...))))))))).))))..)))))))).)))).)))..)))....... ( -36.20) >DroSim_CAF1 37203 101 - 1 GUCCGUGGUGGCCAUGGUAACCAUGAAAUCCAUGAUGUCCAUGAUGCCAAUGGUGUCCGUGGUGUCCAUGAUGCUCAUGGUUUUCCUGGGGAUCCUUACGC ..((((((...))))))...(((.((((.((((((.((.((((((((((.(((...))))))))).))))..)))))))).)))).)))............ ( -34.10) >DroYak_CAF1 37296 92 - 1 GCCAGG---------GGUGUCCAUGAUGUCCAUGGUGUCCAUGAUGCCAAGGGUGUUCGUGAUGUCCAUGGUGUCCGUGGUCUUCAUGGUGUUCCUUACGG ...(((---------..((.((((((.(.((((((........((((((.(((..((...))..))).)))))))))))).).))))))))..)))..... ( -33.60) >consensus GUCCAUGGUGGCCAUGGUAACCAUGAAAUCCAUGAUGUCCAUGAUGCCAAUGGUGUCCGUGGUGUCCAUGAUGCUCAUGGUUUUCCUGGGGAUCCUUACGC ..((((((...))))))...(((.((((.((((((....((((((((((.((.....)))))))).))))....)))))).)))).)))............ (-25.29 = -26.10 + 0.81)


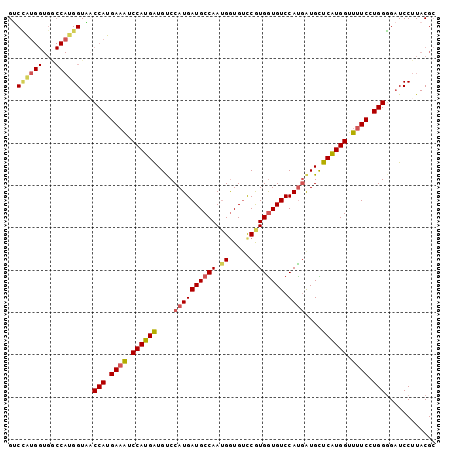
| Location | 9,202,785 – 9,202,905 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -28.69 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202785 120 + 27905053 CCAUGAACAUUGUGGACACCACGGCCAUCAUUGGCAUCAUGGGCAUCAUGGAUUUCAUGGACACCAUGGUCACCAUGGACAUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCA ((((((....(((((...)))))((((....)))).))))))((..(((((..(((((((...((((((...))))))...)))))))..)))))(((......))).......)).... ( -41.30) >DroSec_CAF1 38404 120 + 1 CCAUGAGCAUCAUGGACACCAUGGACACCAUUGGCAUCAUGGACAUCAUGGAUUUCAUGGUUACCAUGGCCACCAUAGCCAUCAUGGACAUCAUGGUCACCAUGGACACUAUUGGCACCA ((((((...((((((...((((((...))).)))..))))))...))))))..((((((((.(((((((...((((.......))))...))))))).)))))))).............. ( -40.10) >DroSim_CAF1 37224 120 + 1 CCAUGAGCAUCAUGGACACCACGGACACCAUUGGCAUCAUGGACAUCAUGGAUUUCAUGGUUACCAUGGCCACCACGGACAUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCA (((((.....))))).......((...(((.(((..((((((..(((((((..((((((((..((.(((...))).))..))))))))..)))))))..))))))...))).)))..)). ( -35.70) >DroYak_CAF1 37317 111 + 1 CCACGGACACCAUGGACAUCACGAACACCCUUGGCAUCAUGGACACCAUGGACAUCAUGGACACC---------CCUGGCAUCAUGGACACCAUGGACAUCAUGGCUACUCUGCGCACCA .(.((((..((((((......(((......)))...((((((...((((((...(((.((....)---------).)))..))))))...))))))...))))))....)))).)..... ( -25.80) >consensus CCAUGAACAUCAUGGACACCACGGACACCAUUGGCAUCAUGGACAUCAUGGAUUUCAUGGACACCAUGGCCACCACGGACAUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCA ((((((...)))))).......((...(((.(((..((((((..(((((((..(((((((...((((((...))))))...)))))))..)))))))..))))))...))).)))..)). (-28.69 = -29.50 + 0.81)



| Location | 9,202,785 – 9,202,905 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -27.12 |
| Energy contribution | -28.88 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202785 120 - 27905053 UGGUGCCAAUAGUGUCCAUGAUGACCAUGAUGUUCAUGAUGUCCAUGGUGACCAUGGUGUCCAUGAAAUCCAUGAUGCCCAUGAUGCCAAUGAUGGCCGUGGUGUCCACAAUGUUCAUGG .((..(((...((..(((((..(((((((.....))))..))))))))..))..)))..))((((((......((((((.(((..((((....))))))))))))).......)))))). ( -36.72) >DroSec_CAF1 38404 120 - 1 UGGUGCCAAUAGUGUCCAUGGUGACCAUGAUGUCCAUGAUGGCUAUGGUGGCCAUGGUAACCAUGAAAUCCAUGAUGUCCAUGAUGCCAAUGGUGUCCAUGGUGUCCAUGAUGCUCAUGG .....(((..((((((.((((..(((((((((..(((.((((.((((((.((....)).))))))....)))).)))..)))((((((...))))))))))))..))))))))))..))) ( -44.60) >DroSim_CAF1 37224 120 - 1 UGGUGCCAAUAGUGUCCAUGAUGACCAUGAUGUUCAUGAUGUCCGUGGUGGCCAUGGUAACCAUGAAAUCCAUGAUGUCCAUGAUGCCAAUGGUGUCCGUGGUGUCCAUGAUGCUCAUGG ...............((((((.(((((((.....))))..)))(((((..(((((((..(((((...(((.(((.....))))))....)))))..)))))))..)))))....)))))) ( -39.00) >DroYak_CAF1 37317 111 - 1 UGGUGCGCAGAGUAGCCAUGAUGUCCAUGGUGUCCAUGAUGCCAGG---------GGUGUCCAUGAUGUCCAUGGUGUCCAUGAUGCCAAGGGUGUUCGUGAUGUCCAUGGUGUCCGUGG ...(((.....)))((((((.....))))))..(((((((((((.(---------((..((.(((((..((.(((((((...))))))).))..).))))))..))).)))))).))))) ( -43.40) >consensus UGGUGCCAAUAGUGUCCAUGAUGACCAUGAUGUCCAUGAUGUCCAUGGUGGCCAUGGUAACCAUGAAAUCCAUGAUGUCCAUGAUGCCAAUGGUGUCCGUGGUGUCCAUGAUGCUCAUGG ((((((.....))).)))......((((((...(((((....((((((..(((((((((((.(((..(((...)))...))))))))).)))))..))))))....)))))...)))))) (-27.12 = -28.88 + 1.75)

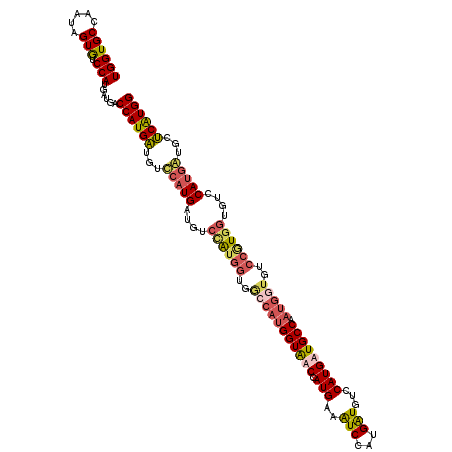

| Location | 9,202,825 – 9,202,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -27.10 |
| Energy contribution | -28.23 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202825 120 + 27905053 GGGCAUCAUGGAUUUCAUGGACACCAUGGUCACCAUGGACAUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAU ((....(((((..(((((((...((((((...))))))...)))))))..)))))(((...((((...(....)...))))....)))...(((((......)))))...))........ ( -35.50) >DroSec_CAF1 38444 120 + 1 GGACAUCAUGGAUUUCAUGGUUACCAUGGCCACCAUAGCCAUCAUGGACAUCAUGGUCACCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAU .......((((..((((((((.(((((((...((((.......))))...))))))).)))))))).....((((..((.....)).....(((((......)))))...))))..)))) ( -38.60) >DroSim_CAF1 37264 120 + 1 GGACAUCAUGGAUUUCAUGGUUACCAUGGCCACCACGGACAUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAU ((....(((((..((((((((..((.(((...))).))..))))))))..)))))(((...((((...(....)...))))....)))...(((((......)))))...))........ ( -32.40) >DroYak_CAF1 37357 111 + 1 GGACACCAUGGACAUCAUGGACACC---------CCUGGCAUCAUGGACACCAUGGACAUCAUGGCUACUCUGCGCACCAUUUCGGUCAUCGUUGGAAUCACCCCGCAUACCAACAUCCU (((..(((((.....))))).....---------..((((...((((....((((.....))))((......))...))))....))))..(((((.((........)).))))).))). ( -24.70) >consensus GGACAUCAUGGAUUUCAUGGACACCAUGGCCACCACGGACAUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAU ((....(((((..(((((((...((((((...))))))...)))))))..)))))(((...((((...(....)...))))....)))...(((((......)))))...))........ (-27.10 = -28.23 + 1.12)


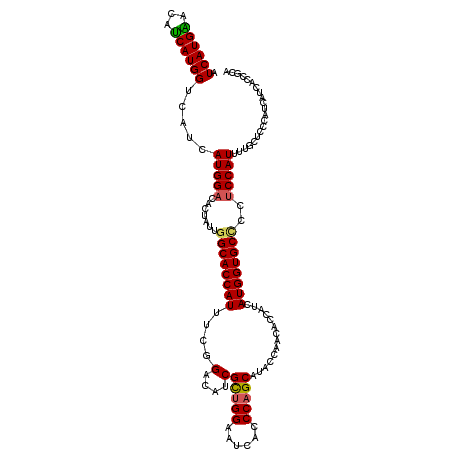
| Location | 9,202,865 – 9,202,982 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202865 117 + 27905053 AUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAUCAUGGUGCCCCUCCAUUUUGCUCCCAUCAUCACCGCA ((((((.....))))))((..(((((.......((((((((...((.....(((((......))))).........))...))))))))..)))))..))................. ( -28.74) >DroSec_CAF1 38484 117 + 1 AUCAUGGACAUCAUGGUCACCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAUCAUGGUGCUCCUCCAUUUUGCUCCCAUCAUCAUCGCA .....(((((..((((.((((((((......((((..((.....)).....(((((......)))))...)))).....)))))))).....))))..)).)))............. ( -30.80) >DroSim_CAF1 37304 117 + 1 AUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAUCAUGGUGCUCCUCCAUUUGGCUGCCAUCAUCAUCGCA ..((((.....))))(((......))).....((((((((..(.(((....(((((......))))).......((((.....))))....))))..))).)))))........... ( -33.10) >DroYak_CAF1 37388 117 + 1 AUCAUGGACACCAUGGACAUCAUGGCUACUCUGCGCACCAUUUCGGUCAUCGUUGGAAUCACCCCGCAUACCAACAUCCUCAUGGUGCCCCUCCAUUUUGCUCCCAUCAUCACCGAA .....(((((..(((((.......((......))(((((((...((.....(((((.((........)).)))))..))..)))))))...)))))..)).)))............. ( -26.20) >consensus AUCAUGAACAUCAUGGUCAUCAUGGACACUAUUGGCACCAUUUCGGACAUCGCUGGAAUCACCCAGCAUACCAACACCAUCAUGGUGCCCCUCCAUUUUGCUCCCAUCAUCACCGCA .((((((...)))))).....(((((.......((((((((....(....)(((((......)))))..............))))))))..)))))..................... (-22.70 = -22.70 + -0.00)


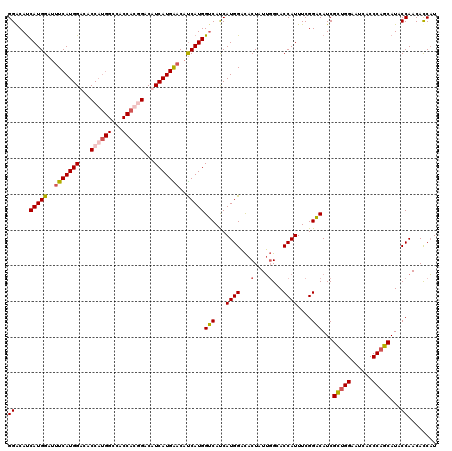
| Location | 9,202,865 – 9,202,982 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -31.39 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9202865 117 - 27905053 UGCGGUGAUGAUGGGAGCAAAAUGGAGGGGCACCAUGAUGGUGUUGGUAUGCUGGGUGAUUCCAGCGAUGUCCGAAAUGGUGCCAAUAGUGUCCAUGAUGACCAUGAUGUUCAUGAU ...(((.((.((((..(((...((....((((((((.......(((((((((((((....)))))).))).)))).))))))))..)).))))))).)).)))(((.....)))... ( -41.00) >DroSec_CAF1 38484 117 - 1 UGCGAUGAUGAUGGGAGCAAAAUGGAGGAGCACCAUGAUGGUGUUGGUAUGCUGGGUGAUUCCAGCGAUGUCCGAAAUGGUGCCAAUAGUGUCCAUGGUGACCAUGAUGUCCAUGAU .......((.((((..(((..((((.....((((((((((.(((((((((((((((....)))))))....((.....)))))))))).))).))))))).))))..))))))).)) ( -41.40) >DroSim_CAF1 37304 117 - 1 UGCGAUGAUGAUGGCAGCCAAAUGGAGGAGCACCAUGAUGGUGUUGGUAUGCUGGGUGAUUCCAGCGAUGUCCGAAAUGGUGCCAAUAGUGUCCAUGAUGACCAUGAUGUUCAUGAU ((.(((..((.(((((.(((..((((.(((((((.....))))))....(((((((....))))))).).))))...)))))))).))..))))).......((((.....)))).. ( -38.10) >DroYak_CAF1 37388 117 - 1 UUCGGUGAUGAUGGGAGCAAAAUGGAGGGGCACCAUGAGGAUGUUGGUAUGCGGGGUGAUUCCAACGAUGACCGAAAUGGUGCGCAGAGUAGCCAUGAUGUCCAUGGUGUCCAUGAU (((.((..((.......))..)).)))(((((((((..((((((((((.((..(((....)))..))...)))...(((((((.....))).))))))))))))))))))))..... ( -35.40) >consensus UGCGAUGAUGAUGGGAGCAAAAUGGAGGAGCACCAUGAUGGUGUUGGUAUGCUGGGUGAUUCCAGCGAUGUCCGAAAUGGUGCCAAUAGUGUCCAUGAUGACCAUGAUGUCCAUGAU ...((..((.((((...((..(((((.(.(((((((.......(((((((((((((....)))))).))).)))).)))))))......).)))))..)).)))).))..))..... (-31.39 = -32.20 + 0.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:04 2006