| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,197,358 – 9,197,448 |
| Length | 90 |
| Max. P | 0.964912 |

| Location | 9,197,358 – 9,197,448 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.95 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
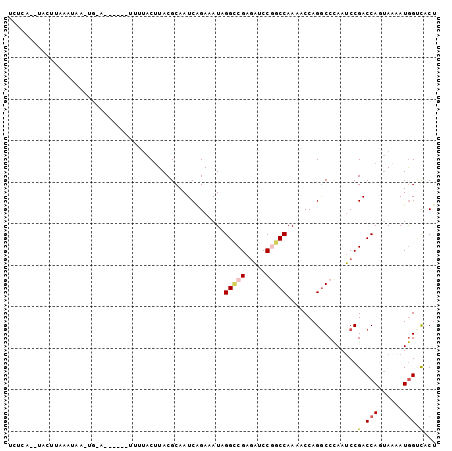
>3R_DroMel_CAF1 9197358 90 + 27905053 UUUCAAACUCUUAAA-------U------UUUUACUUACGCAAUCAGAAAUAGGCCGAGAUCCGGCCAAAACCAGGCCCAAUCCGACCAGUAAAAUGGUCACU ...............-------.------.......................(((((.....))))).......((......))(((((......)))))... ( -18.20) >DroSec_CAF1 34488 97 + 1 UCUCAAAUACUUAACUAAAUGAG------UGUUACUUACGCAAUCAGAAAUAGGUCGAGAUCCUGCCAAAACCAGGCCCAAUCCGACCAUUAUGAUGGUCACU .....((((((((......))))------))))......((.((((..(((.(((((.(((...(((.......)))...))))))))))).)))).)).... ( -22.90) >DroSim_CAF1 33304 97 + 1 UCUCAAAUACUUAACUAAAUGAG------UGUUACUUACGCAAUCAGAAAUAGGCCGAGAUCCGGCCAAAACCAGGCCCAAUCCGACCAGUAAAAUGGUCACU ((((.((((((((......))))------))))......((....(....)..)).))))...((((.......))))......(((((......)))))... ( -23.50) >DroEre_CAF1 40041 95 + 1 -------UACUUGAAUAAGUGUAUAACU-UUAUACUUACGCAAUCGGAAAUAGGCCGAGACCCGGCCAAAACCAGACCCAAUCCGACCAGUAAAAUGGCCGCU -------...(((..((((((((.....-.))))))))..)))(((((....(((((.....))))).......(...)..)))))(((......)))..... ( -23.80) >DroYak_CAF1 33308 101 + 1 ACGCC--AGCUUGAAUAAGUGUAUAGCUUUUAUACGUACGCAAUCGGAAAUAGGCCGAGACCCGGCCAAAACCAGGCCCAAUCCGACCAAUAAAAUGGCCACU ..(((--(..(((.....((((((...........))))))..(((((....(((((.....)((......)).))))...)))))....)))..)))).... ( -24.40) >DroAna_CAF1 36409 71 + 1 --------------------------------GACUUACGCAAUCCGAGAUGGGGCGGCUGCCAGCCAAAACCAGGCCUAAACCGACCAGAAAAAUAGAUGCU --------------------------------.......(((.((.....(((..(((......(((.......))).....))).)))........))))). ( -12.72) >consensus UCUCA__UACUUAAAUAA_UG_A______UUUUACUUACGCAAUCAGAAAUAGGCCGAGAUCCGGCCAAAACCAGGCCCAAUCCGACCAGUAAAAUGGUCACU ....................................................(((((.....))))).................(.(((......))).)... ( -8.53 = -8.95 + 0.42)


| Location | 9,197,358 – 9,197,448 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
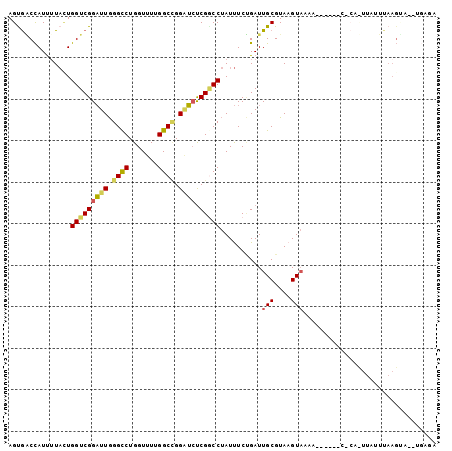
>3R_DroMel_CAF1 9197358 90 - 27905053 AGUGACCAUUUUACUGGUCGGAUUGGGCCUGGUUUUGGCCGGAUCUCGGCCUAUUUCUGAUUGCGUAAGUAAAA------A-------UUUAAGAGUUUGAAA (((((.....)))))(((((((((.((((.......)))).)))).))))).........((((....))))..------.-------............... ( -27.30) >DroSec_CAF1 34488 97 - 1 AGUGACCAUCAUAAUGGUCGGAUUGGGCCUGGUUUUGGCAGGAUCUCGACCUAUUUCUGAUUGCGUAAGUAACA------CUCAUUUAGUUAAGUAUUUGAGA ........(((.((.(((((((((..(((.......)))..)))).)))))...)).)))((((....))))..------((((..((......))..)))). ( -26.10) >DroSim_CAF1 33304 97 - 1 AGUGACCAUUUUACUGGUCGGAUUGGGCCUGGUUUUGGCCGGAUCUCGGCCUAUUUCUGAUUGCGUAAGUAACA------CUCAUUUAGUUAAGUAUUUGAGA (((((.....)))))(((((((((.((((.......)))).)))).)))))......((.((((....))))))------((((..((......))..)))). ( -31.00) >DroEre_CAF1 40041 95 - 1 AGCGGCCAUUUUACUGGUCGGAUUGGGUCUGGUUUUGGCCGGGUCUCGGCCUAUUUCCGAUUGCGUAAGUAUAA-AGUUAUACACUUAUUCAAGUA------- ..((((((......))))))((.(((((.(((....(((((.....))))).....))).........(((((.-...)))))))))).)).....------- ( -26.30) >DroYak_CAF1 33308 101 - 1 AGUGGCCAUUUUAUUGGUCGGAUUGGGCCUGGUUUUGGCCGGGUCUCGGCCUAUUUCCGAUUGCGUACGUAUAAAAGCUAUACACUUAUUCAAGCU--GGCGU ....((((.(((...(((((((((.((((.......)))).)))).))))).................(((((.....)))))........))).)--))).. ( -30.80) >DroAna_CAF1 36409 71 - 1 AGCAUCUAUUUUUCUGGUCGGUUUAGGCCUGGUUUUGGCUGGCAGCCGCCCCAUCUCGGAUUGCGUAAGUC-------------------------------- .((((((........((.(((((..((((.......))))...))))))).......))).))).......-------------------------------- ( -19.06) >consensus AGUGACCAUUUUACUGGUCGGAUUGGGCCUGGUUUUGGCCGGAUCUCGGCCUAUUUCUGAUUGCGUAAGUAAAA______C_CA_UUAUUUAAGUA__UGAGA ...............(((((((((.((((.......)))).)))).)))))..........(((....)))................................ (-19.85 = -19.88 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:56 2006