
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,163,721 – 9,163,818 |
| Length | 97 |
| Max. P | 0.995315 |

| Location | 9,163,721 – 9,163,818 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.87 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.01 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9163721 97 + 27905053 AAGA--ACCGCUGUUAGUUAACA----------UUUGG------CUAACAGAAAC-C-GUCGACAAGUGCCACGAUUGCCGCAAGGCAACUCAGUCUUUCACGGACUUUCCUUCAAA ..((--(...((((((((((...----------..)))------)))))))...(-(-((.((.(((.((...(((((((....))))).)).))))))))))).......)))... ( -28.20) >DroPse_CAF1 55428 109 + 1 AAGGCGAGAGCUGUUGGUUAACAGAGCUCGCGCUGUGG------CUAACAGCAGA-C-GCGAAGUGCUGCCUCAAUUGCCGCAAGGCAACUCAGUCUUCAACGGACUACGCUGAACU ..((((((.(((((((((((.(((.(....).))))))------)))))))).((-.-(((......))).))..(((((....))))))))(((((.....)))))..)))..... ( -42.20) >DroYak_CAF1 57006 97 + 1 AAGA--ACCGCUGUUAGUUAACA----------UUUGG------CUAACAGAAGC-C-GUCGACAAGUGCCACGAUUGCCGCAAGGCAACUCAGUCUUUCACCGACUUUCCUUCAUU ..((--(...((((((((((...----------..)))------)))))))....-.-((((....(((..(((((((((....))))).)).))....))))))).....)))... ( -26.70) >DroMoj_CAF1 75039 105 + 1 GCGAC-GCCACUGUUAGCUAACA----------GCUGUUAACCCUUAACAGCCCAGC-GCUAACUGCCGUUGAAAUUGCCACACGGAAAUUCAGUCUAACAUUGACUGCGCUGCCAA .((((-(.((..((((((.....----------(((((((.....))))))).....-)))))))).)))))............((.....(((((.......))))).....)).. ( -29.90) >DroAna_CAF1 57394 98 + 1 AAGA--AGUGCUGUUGGUUAACA----------GGCGG------CUAACAGCAGG-CAGCCAAACGAUGCCACGAUUGCCGCAAGGCAACUCAGUCUGUCAACGACUUCGCUCAAAU ..((--(((((((((((((....----------...))------)))))))).((-(((.(...((......)).(((((....)))))....).)))))....)))))........ ( -34.70) >DroPer_CAF1 64997 109 + 1 AAGGCGAGAGCUGUUGGUUAACAGAGCUCGCGCUGUGU------CUAACAGCAGA-C-GCGAAGUGCUGCCUCAAUUGCCGCAAGGCAACUCAGUCUUCAACGGACUACGCUGAACU ..((((((..(((((....)))))..)))(((((((((------((......)))-)-))..))))).)))(((.(((((....)))))...(((((.....)))))....)))... ( -42.10) >consensus AAGA__ACCGCUGUUAGUUAACA__________UGUGG______CUAACAGCAGA_C_GCCAACUGCUGCCACAAUUGCCGCAAGGCAACUCAGUCUUUCACGGACUUCGCUGAAAU ..........(((((((...........................)))))))........................(((((....)))))...((((.......)))).......... (-15.23 = -15.01 + -0.22)

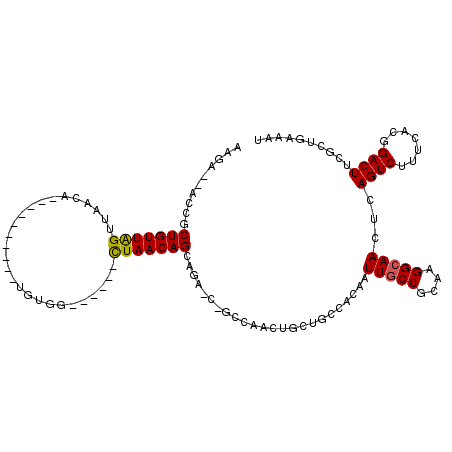

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:45 2006