
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,157,305 – 9,157,398 |
| Length | 93 |
| Max. P | 0.522579 |

| Location | 9,157,305 – 9,157,398 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9157305 93 - 27905053 UUGCU----A----------UUUUGCUAUUUAUGU---UCUUAGGAAUGCCGCAGGACCUCUAUAACCUCUGGCACCCGGAUAAUUAUCCUUUCAGUGACAGCAUCUGCA .(((.----.----------...((((..((((..---.....((..(((((.(((..........))).))))).))((((....)))).....)))).))))...))) ( -19.90) >DroGri_CAF1 73285 104 - 1 UUCCCCCCAAUUUAC--UAACUGUGGAU-CUGU---GCUCGUAGGCAUGCCACAGGAUCUGUACAACUUGUGGCACCCGGACAACUAUCCGUUCUCUGACGGCAUUUGCA .........((((((--.....))))))-..((---((.(((.((..(((((((((..........)))))))))))((((.(((.....))).)))))))))))..... ( -27.20) >DroSec_CAF1 48314 103 - 1 UUGCC----AUUUAUUCGAAUUAUGCGUCUUAUGU---UCUUAGGAAUGCCGCAGGACCUCUAUAACCUCUGGCACCCGGAUAAUUAUCCUUUCAGUGACAGCAUCUGCA .....----.............(((((((...((.---.....((..(((((.(((..........))).))))).))((((....))))...))..))).))))..... ( -20.40) >DroSim_CAF1 43280 97 - 1 UUGCU----CUUUA------UUAUGCGUCUUAUGU---UCUUAGGAAUGCCGCAGGACCUCUAUAACCUCUGGCACCCGGAUAAUUAUCCUUUCAGUGACAGCAUCUGUA .....----.....------..(((((((...((.---.....((..(((((.(((..........))).))))).))((((....))))...))..))).))))..... ( -20.40) >DroEre_CAF1 53795 84 - 1 --------------------------GUCUUAUAUUCCUCUUAGGAAUGCCGCAGGACCUCUAUAACCUCUGGCACCCGGAUAAUUAUCCCUUCAGCGACAGCAUCUGCA --------------------------(((....(((((.....)))))((((.(((..........))).))))....((((....)))).......))).((....)). ( -19.40) >DroYak_CAF1 50197 103 - 1 ---CU----AUUUAUUCGUGCUAUACGUCUUAUAUUCUUCUUAGGAAUGCCGCAGGACCUCUAUAACCUCUGGCACCCGGAUAAUUAUCCCUUUAGCGACAGCAUCUGCA ---..----........((((((....................((..(((((.(((..........))).))))).))((((....))))...)))).)).((....)). ( -19.90) >consensus UUGCU____AUUUA______UUAUGCGUCUUAUGU___UCUUAGGAAUGCCGCAGGACCUCUAUAACCUCUGGCACCCGGAUAAUUAUCCUUUCAGUGACAGCAUCUGCA ...........................................((..(((((.(((..........))).))))).))((((....))))...........((....)). (-14.50 = -14.25 + -0.25)

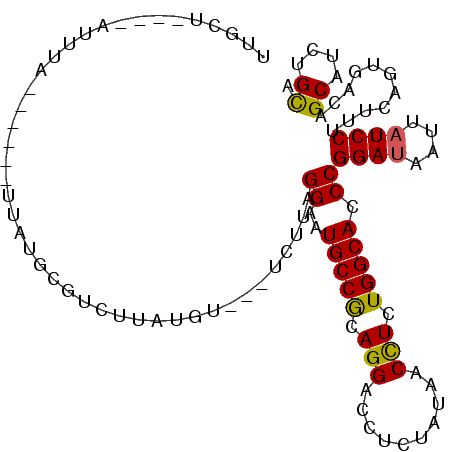

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:43 2006