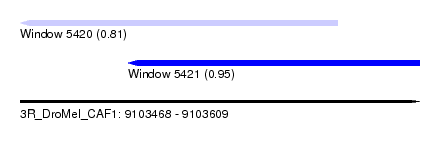
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,103,468 – 9,103,609 |
| Length | 141 |
| Max. P | 0.949814 |

| Location | 9,103,468 – 9,103,580 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
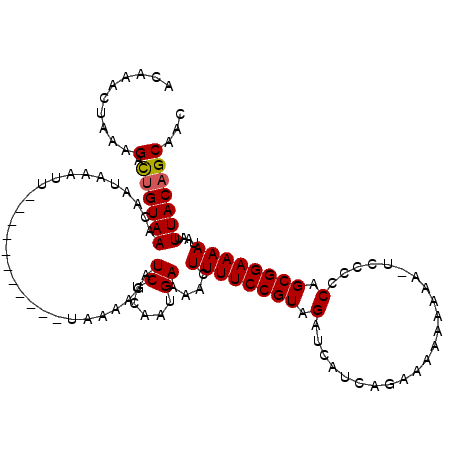
>3R_DroMel_CAF1 9103468 112 - 27905053 AAAAGAUCUCAAUGAAAACUUUUCCGUAGAUCAUCAAAAAAAAAAA-A-UCCCAGCGGAAAAUAAUUUACAGCAUUUAC--AUUUUUUUCCAUGGCUGGCUGGCUUGUCAACCAGA ....(((((....(((.....)))...)))))..............-.-..(((((((((((.(((.((.......)).--))).))))))...)))))((((........)))). ( -20.00) >DroSim_CAF1 14332 112 - 1 AAAAGAUCUCAAUGAAAACUUUUCCGUAGAUCAUCAGCUAAAAAAA-UCCCCCAGCGGAAAAUAAAUUACAGCACCACCUGAUUUUUUUCCAUA---AGCUGGCUGGUCAACCAGA ....(((((....(((.....)))...)))))..............-....(((((((((((.((((..(((......)))))))))))))...---.)))))((((....)))). ( -22.20) >DroYak_CAF1 13534 112 - 1 AAAUGAUCUCAACGAGAACUUUUCCGUAGAUCAUCGGGAAAAAAAAAUCCCCCAGCGGAAAAUAGUUUACAGCAACACCUGAU-UUUUUCCAUA---AGCUGGCUGGUCAACCGGA ..(((((((..(((.((.....)))))))))))).((((........))))(((((((((((.......(((......)))..-.))))))...---.)))))((((....)))). ( -29.10) >consensus AAAAGAUCUCAAUGAAAACUUUUCCGUAGAUCAUCAGAAAAAAAAA_UCCCCCAGCGGAAAAUAAUUUACAGCAACACCUGAUUUUUUUCCAUA___AGCUGGCUGGUCAACCAGA ....(((((....((((...))))...)))))...................(((((((((((.......................)))))).......)))))((((....)))). (-16.75 = -16.64 + -0.11)


| Location | 9,103,506 – 9,103,609 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -16.02 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9103506 103 - 27905053 ACAAACUAAAGACUGUAAACAAUAAAUU-----------CAAAAGAUCUCAAUGAAAACUUUUCCGUAGAUCAUCAAAAAAAAAAA-A-UCCCAGCGGAAAAUAAUUUACAGCAUU ..........(.(((((((.......((-----------((...........))))...((((((((.(((...............-)-))...))))))))...))))))))... ( -13.26) >DroSim_CAF1 14369 102 - 1 --AAAACAAAGACCGUAAACAAUAAAUA-----------UAAAAGAUCUCAAUGAAAACUUUUCCGUAGAUCAUCAGCUAAAAAAA-UCCCCCAGCGGAAAAUAAAUUACAGCACC --............((((..........-----------.....(((((....(((.....)))...)))))..............-(((......))).......))))...... ( -8.70) >DroYak_CAF1 13570 116 - 1 ACAAACUGAAGGUUGUAAACAAUAAAUUUAUAUUGUAAAUAAAUGAUCUCAACGAGAACUUUUCCGUAGAUCAUCGGGAAAAAAAAAUCCCCCAGCGGAAAAUAGUUUACAGCAAC ...........(((((((((.....((((((.......))))))..((((...))))..((((((((.(.....)((((........))))...))))))))..)))))))))... ( -26.10) >consensus ACAAACUAAAGACUGUAAACAAUAAAUU___________UAAAAGAUCUCAAUGAAAACUUUUCCGUAGAUCAUCAGAAAAAAAAA_UCCCCCAGCGGAAAAUAAUUUACAGCAAC ..........(.((((((............................((.....))....((((((((.(.......................).))))))))....)))))))... ( -9.42 = -9.53 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:26 2006