
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,099,360 – 9,099,498 |
| Length | 138 |
| Max. P | 0.595185 |

| Location | 9,099,360 – 9,099,458 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -20.01 |
| Energy contribution | -21.38 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9099360 98 + 27905053 ---CCCUCCAUCCAACGAAAAGCUCACGAAAACGGUCGAAAUGCGAACGAGUUUUGGCCGCUUAUCAA--------UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU-----------GA ---.......((.(((((....((((.(....((((((((((.(....).))))))))))....((((--------..((....))..))))).))))...))))).-----------)) ( -24.50) >DroSec_CAF1 9066 104 + 1 ---CCCUCCAUCCAACGAAAAGCUCACGAAAACGGUCGAAAUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCAA--UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU-----------GA ---.......................(((......))).....(((((((((((((((((.....(((.(((..--..))).)))....)))))..)))))))))))-----------). ( -27.70) >DroSim_CAF1 10228 104 + 1 ---CCCUCCAUCCAACGAAAAGCUCACGAAAACGGUCGAAAUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCAA--UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU-----------GA ---.......................(((......))).....(((((((((((((((((.....(((.(((..--..))).)))....)))))..)))))))))))-----------). ( -27.70) >DroEre_CAF1 9992 107 + 1 ----CCGCCA-CCAACGAAAAGCUCACGAAAACGGUCGAAAUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCUU--UUG------ACUUUGGCCUGAGAACUCGUUUGCUUUGCGCCUGA ----......-.....................(((.((....((((((((((((((((((....((((......--)))------)...)))))..)))))))))))))....)).))). ( -32.80) >DroYak_CAF1 9323 114 + 1 ---CCCGCCA-CCAACGAAAAGCUCACGAAAACGGUCGAAAUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCUU--UUGCUAUUGCUUUUGGCCUGAGAACUCGUUUGCUUUGCGCCUGA ---.......-.....................(((.((....((((((((((((((((((.....(((((((..--..)))))))....)))))..)))))))))))))....)).))). ( -39.50) >DroAna_CAF1 12569 115 + 1 UGGCUCUACAUCCCCC----ACCUCGUGC-AACGGUCCAAAUCCGAACGAGUUUUGGCCGCUUAUCAAUAGCGAUAUAGCCGUUGCCUUUGGCCCGAGAACUCGUUUGCUUGGCGGCUGA .((((.((......((----(.(((((..-..(((.......))).)))))...))).((((.......)))).)).))))((((((...(((.(((....)))...))).))))))... ( -34.00) >consensus ___CCCUCCAUCCAACGAAAAGCUCACGAAAACGGUCGAAAUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCAA__UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU___________GA .....................((...(((......)))....))((((((((((((((((((.......)))......((....))....))))..)))))))))))............. (-20.01 = -21.38 + 1.38)

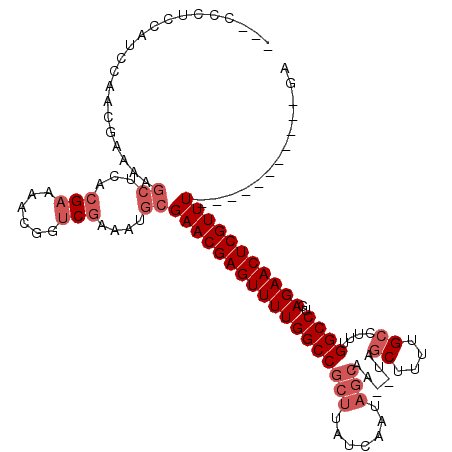

| Location | 9,099,397 – 9,099,498 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -26.39 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9099397 101 + 27905053 AUGCGAACGAGUUUUGGCCGCUUAUCAA--------UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU-----------GACUUUGGGCCCUGUUACAAUUUGCGUUUGCCUGCUUAGCAU ((((((((((((((((((((.....(((--------......)))....)))))..)))))))))))-----------......((((..(((........)))...)))).....)))) ( -28.70) >DroSec_CAF1 9103 107 + 1 AUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCAA--UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU-----------GACUUUGGGCCCUGUUACAAUUUGCGUUUGCCUGCUUAGCAU ((((((((((((((((((((.....(((.(((..--..))).)))....)))))..)))))))))))-----------......((((..(((........)))...)))).....)))) ( -32.90) >DroSim_CAF1 10265 107 + 1 AUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCAA--UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU-----------GACUUUGGGCCCUGUUACAAUUUGCGUUUGCCUGCUUAGCAU ((((((((((((((((((((.....(((.(((..--..))).)))....)))))..)))))))))))-----------......((((..(((........)))...)))).....)))) ( -32.90) >DroEre_CAF1 10027 112 + 1 AUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCUU--UUG------ACUUUGGCCUGAGAACUCGUUUGCUUUGCGCCUGACUUUGGGCCCUGUUACAAUUUGCCUUUGCCUGCUUAGCAU ..((((((((((((((((((....((((......--)))------)...)))))..)))))))))))))..(((((((......)))).............((........))...))). ( -35.50) >DroYak_CAF1 9359 118 + 1 AUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCUU--UUGCUAUUGCUUUUGGCCUGAGAACUCGUUUGCUUUGCGCCUGACUUUGGGCCCUGUUACAAUUUGCCUUUGCCUGCUUAGCAU ..((((((((((((((((((.....(((((((..--..)))))))....)))))..)))))))))))))..(((((((......)))).............((........))...))). ( -42.20) >DroAna_CAF1 12604 120 + 1 AUCCGAACGAGUUUUGGCCGCUUAUCAAUAGCGAUAUAGCCGUUGCCUUUGGCCCGAGAACUCGUUUGCUUGGCGGCUGACUUUGGGCCCUGUUACAAUUUGCUUUUGCCUGCUUUGCAU ....((((((((((((((((.....(((..((......))..)))....)))))..)))))))))))((..(((((((.......))))............((....))..)))..)).. ( -37.30) >consensus AUGCGAACGAGUUUUGGCCGCUUAUCAAUAGCAA__UAGCUUUUGCCUUUGGCCUGAGAACUCGUUU___________GACUUUGGGCCCUGUUACAAUUUGCGUUUGCCUGCUUAGCAU ((((((((((((((((((((((.......)))......((....))....))))..))))))))))).................((((...((........))....)))).....)))) (-26.39 = -27.27 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:23 2006