
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
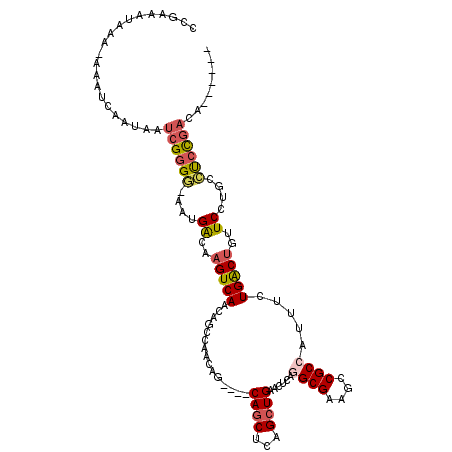
| Location | 9,077,408 – 9,077,512 |
| Length | 104 |
| Max. P | 0.764469 |

| Location | 9,077,408 – 9,077,512 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -16.03 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
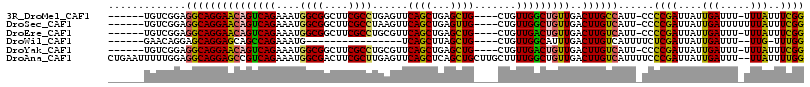
>3R_DroMel_CAF1 9077408 104 + 27905053 CCGAAAUAAA-AAAUCAAUAAUCGGGG-AAUGGCAAGUCAACAGCCAACAG----CAGCUCAGCUGAACUCAGGCGAAGCCGCCAUUUCUGACUGUUCCUGCCUCCGACA------ ..........-..........((((((-...((..(((((........(((----(......))))......((((....)))).....)))))...))...))))))..------ ( -28.90) >DroSec_CAF1 191710 105 + 1 CCGAAAUAAAAAAAUCAAUAAUCGGGG-AAUGACAAGUCAACAGCCAACAG----CAACUCAGCUGAACUUAGGCGAAGCCGCCAUUUCUGACUGUUCCUGCCUCCGACA------ .....................((((((-...((((.((((........(((----(......))))......((((....)))).....))))))))....)).))))..------ ( -27.50) >DroEre_CAF1 198165 104 + 1 CCGAAAUAAA-AAAUCAAUAAUCGGGG-AAUGACAAGUCAACAGUCAACAG----CAGCUCAGCUGAACGCAGGCGAAGCCGCCAUUUCUGACUGUUCCUGCCUCCGACA------ ..........-..........((((((-..(((....)))(((((((.(((----(......))))......((((....)))).....)))))))......))))))..------ ( -30.80) >DroWil_CAF1 197995 87 + 1 CCAAA-CAA--AAAUCAAUAAUCGAGAAAAUGACAAGUCAAAUGCCAACAG----CAGCUAAGCUGA----------------CAUUUCUGGCUGCUCCUGCUCCUGUUC------ .....-...--............(((....(((....))).....((..((----((((((......----------------......))))))))..)))))......------ ( -13.50) >DroYak_CAF1 207015 104 + 1 CCGAAAUAAA-AAAUCAAUAAUCGGGG-AAUGACAAGUCAACAGUCAACAG----CAGCUCAGCUGAACGCAGGCGAAGCCGCCAUUUCUGACUGUUCCUGCCUCCGACA------ ..........-..........((((((-..(((....)))(((((((.(((----(......))))......((((....)))).....)))))))......))))))..------ ( -30.80) >DroAna_CAF1 203369 114 + 1 CCAAAAUAA--AAAUCAAUAAUCGGGAAAAUGACAAGUCAACAGCCAAAAGCAAGCAGCUGAGCUGAACUCAAGCGAAGUCGCCAUUUCUGACGGCUCCUGCCUCCAAAAAUUCAG .........--.............(((.........((((.((((....(((.....)))..)))).......(((....)))......))))(((....)))))).......... ( -23.50) >consensus CCGAAAUAAA_AAAUCAAUAAUCGGGG_AAUGACAAGUCAACAGCCAACAG____CAGCUCAGCUGAACUCAGGCGAAGCCGCCAUUUCUGACUGUUCCUGCCUCCGACA______ .....................((((((....((..(((((...............((((...))))......((((....)))).....)))))..))....))))))........ (-16.03 = -16.72 + 0.68)


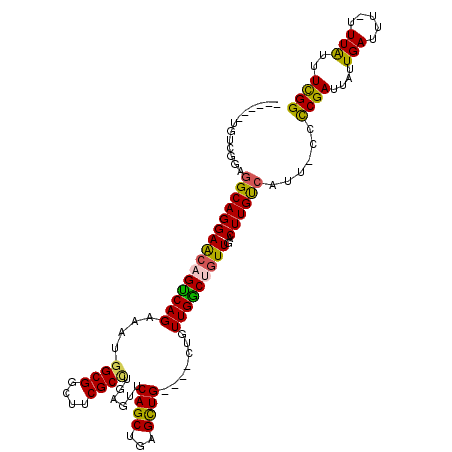
| Location | 9,077,408 – 9,077,512 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -23.23 |
| Energy contribution | -22.72 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9077408 104 - 27905053 ------UGUCGGAGGCAGGAACAGUCAGAAAUGGCGGCUUCGCCUGAGUUCAGCUGAGCUG----CUGUUGGCUGUUGACUUGCCAUU-CCCCGAUUAUUGAUUU-UUUAUUUCGG ------....((((((((((((((((((....((((((((.((.((....)))).))))))----)).)))))))))..))))))..)-))((((....(((...-.)))..)))) ( -40.30) >DroSec_CAF1 191710 105 - 1 ------UGUCGGAGGCAGGAACAGUCAGAAAUGGCGGCUUCGCCUAAGUUCAGCUGAGUUG----CUGUUGGCUGUUGACUUGUCAUU-CCCCGAUUAUUGAUUUUUUUAUUUCGG ------....((((((((((((((((((....((((....))))......((((......)----))))))))))))..))))))..)-))((((....(((.....)))..)))) ( -32.90) >DroEre_CAF1 198165 104 - 1 ------UGUCGGAGGCAGGAACAGUCAGAAAUGGCGGCUUCGCCUGCGUUCAGCUGAGCUG----CUGUUGACUGUUGACUUGUCAUU-CCCCGAUUAUUGAUUU-UUUAUUUCGG ------....((((((((((((((((((....((((((((.((.((....)))).))))))----)).)))))))))..))))))..)-))((((....(((...-.)))..)))) ( -36.60) >DroWil_CAF1 197995 87 - 1 ------GAACAGGAGCAGGAGCAGCCAGAAAUG----------------UCAGCUUAGCUG----CUGUUGGCAUUUGACUUGUCAUUUUCUCGAUUAUUGAUUU--UUG-UUUGG ------((((((((((((.((((((((....))----------------...)))..))).----))))(((((.......))))).................))--)))-))).. ( -20.40) >DroYak_CAF1 207015 104 - 1 ------UGUCGGAGGCAGGAACAGUCAGAAAUGGCGGCUUCGCCUGCGUUCAGCUGAGCUG----CUGUUGACUGUUGACUUGUCAUU-CCCCGAUUAUUGAUUU-UUUAUUUCGG ------....((((((((((((((((((....((((((((.((.((....)))).))))))----)).)))))))))..))))))..)-))((((....(((...-.)))..)))) ( -36.60) >DroAna_CAF1 203369 114 - 1 CUGAAUUUUUGGAGGCAGGAGCCGUCAGAAAUGGCGACUUCGCUUGAGUUCAGCUCAGCUGCUUGCUUUUGGCUGUUGACUUGUCAUUUUCCCGAUUAUUGAUUU--UUAUUUUGG ...(((..((((((((.((((.(((((....))))).))))))))((((((((((.(((.....)))...)))))..))))).........))))..))).....--......... ( -31.90) >consensus ______UGUCGGAGGCAGGAACAGUCAGAAAUGGCGGCUUCGCCUGAGUUCAGCUGAGCUG____CUGUUGGCUGUUGACUUGUCAUU_CCCCGAUUAUUGAUUU_UUUAUUUCGG .............(((((((((((((((....((((....))))......((((...)))).......)))))))))..))))))......((((....(((.....)))..)))) (-23.23 = -22.72 + -0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:15 2006