| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,070,083 – 9,070,180 |
| Length | 97 |
| Max. P | 0.860897 |

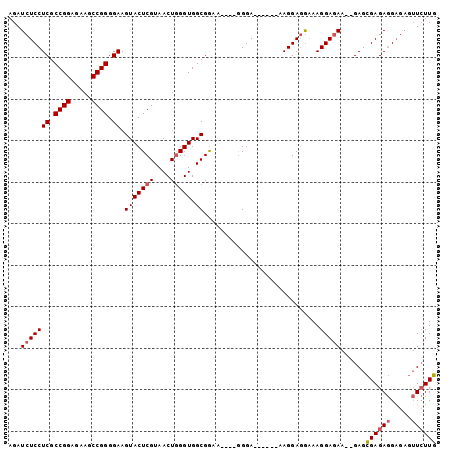
| Location | 9,070,083 – 9,070,180 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9070083 97 + 27905053 AGAUCUCCUCGCCGGAGAAGCCGGGGAAGUACUCGUAACUAGGUGGCGGAA----GGGAGGAAGGGAGGAGGAAAGGAAAA--GAGCGAGAGGAGAGUUCUUG ...(((((((.((((.....)))).......(((.(..((......(....----)......))..).)))..........--......)))))))....... ( -25.30) >DroSec_CAF1 183807 91 + 1 AGAUCUCCUCGCCGGAGAAGCCGGGGAAGUACUCGUAACUGGGUGGCGGUG----GGGA------AAGGAGGAAAGGAGAA--GAGCGAGAGGCGAGUUCUUG .....(((((((((.....(((.((.............)).)))..)))))----))))------..............((--((((.........)))))). ( -25.42) >DroSim_CAF1 187623 91 + 1 AGAUCUCCUCGCCGGAGAAGCCGGGGAAGUACUCGUAACUGGGUGGCGGUG----GGGA------AAGGAGGAAAGGAGAA--GAGCGAGAGGAGAGUUCUUG .....(((((((((.....(((.((.............)).)))..)))))----))))------..............((--((((.........)))))). ( -25.42) >DroYak_CAF1 198513 97 + 1 AGAUCUCCUCGCCGGAGAAGCCGGGGAAGUACUCGUAACUGGGUGGCGAAAAAAAAGGA------AAGGAAAGAAGGAGAGGAGAGAGAGAAGAGCGAUGUUU ...(((((((.((((.....))))....(((((((....))))).))............------.............))))))).................. ( -22.30) >consensus AGAUCUCCUCGCCGGAGAAGCCGGGGAAGUACUCGUAACUGGGUGGCGGAA____GGGA______AAGGAGGAAAGGAGAA__GAGCGAGAGGAGAGUUCUUG ...(((((((.((((.....)))).)).(((((((....))))).))............................)))))......((((((.....)))))) (-18.90 = -19.52 + 0.63)


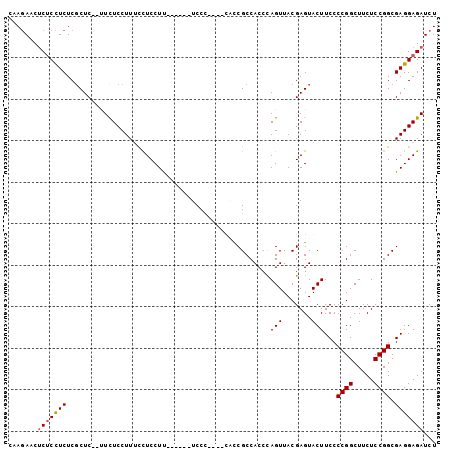
| Location | 9,070,083 – 9,070,180 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -16.73 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9070083 97 - 27905053 CAAGAACUCUCCUCUCGCUC--UUUUCCUUUCCUCCUCCCUUCCUCCC----UUCCGCCACCUAGUUACGAGUACUUCCCCGGCUUCUCCGGCGAGGAGAUCU ..(((..(((((((..((((--..........................----.................))))......((((.....)))).)))))))))) ( -17.53) >DroSec_CAF1 183807 91 - 1 CAAGAACUCGCCUCUCGCUC--UUCUCCUUUCCUCCUU------UCCC----CACCGCCACCCAGUUACGAGUACUUCCCCGGCUUCUCCGGCGAGGAGAUCU ..((((...((.....))..--))))......((((((------.((.----....(((....(((.......))).....)))......)).)))))).... ( -15.00) >DroSim_CAF1 187623 91 - 1 CAAGAACUCUCCUCUCGCUC--UUCUCCUUUCCUCCUU------UCCC----CACCGCCACCCAGUUACGAGUACUUCCCCGGCUUCUCCGGCGAGGAGAUCU ..(((..(((((((..((((--................------....----.................))))......((((.....)))).)))))))))) ( -17.69) >DroYak_CAF1 198513 97 - 1 AAACAUCGCUCUUCUCUCUCUCCUCUCCUUCUUUCCUU------UCCUUUUUUUUCGCCACCCAGUUACGAGUACUUCCCCGGCUUCUCCGGCGAGGAGAUCU ..................(((((((.............------.........((((..((...))..)))).......((((.....)))).)))))))... ( -16.70) >consensus CAAGAACUCUCCUCUCGCUC__UUCUCCUUUCCUCCUU______UCCC____CACCGCCACCCAGUUACGAGUACUUCCCCGGCUUCUCCGGCGAGGAGAUCU .......(((((((.................................................(((.......)))...((((.....)))).)))))))... (-12.59 = -12.90 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:13 2006