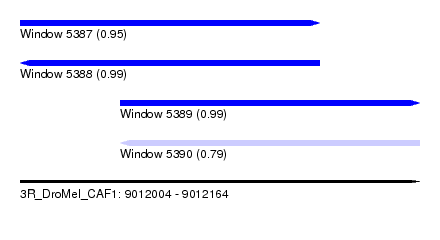
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,012,004 – 9,012,164 |
| Length | 160 |
| Max. P | 0.989760 |

| Location | 9,012,004 – 9,012,124 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -22.50 |
| Energy contribution | -24.50 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9012004 120 + 27905053 CUGGACAAAAGCCUCUGGCUUACCAGCCUCCACAUAAACUUGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCGUUUGAUAGCUCAUUCCGCUCCUGCUGGCUUUCGUCCUCUAUUUC ..((((.((((((...((((....))))............((((((......))))))..((((((.((((....)))).))))))......((....)).)))))).))))........ ( -35.00) >DroSec_CAF1 125979 120 + 1 CUGGACAAAAGCGUCUGGCUUACCAGCCUGCACAUAAACUUGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCAUUUGAUAGCUCCUUCCGCUCCUGCUGGCUUUCGUCCUUUAUUUC ..((((..((((.....)))).(((((..((.........((((((......))))))..((((((.((((....)))).))))))......))....))))).....))))........ ( -34.20) >DroSim_CAF1 129219 120 + 1 CUGGACAAAAGCCUCUGGCUUACCAGCCUGCACAUAAACUUGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCAUUUGAUAGCUCCUUCCGCUCCUGCUGGCUUUCGUCCUUUAUUUC ..((((.((((((...((((....)))).(((........((((((......))))))..((((((.((((....)))).))))))...........))).)))))).))))........ ( -36.40) >DroEre_CAF1 132018 120 + 1 CUGGACAAAAGCCUCCGUCUUACCAGCCUCCACAUAGACUUGCACAUUAUCCAGAGCCUUGGCUAUGCCCCUUUCCUUCGAUGGCUCCUUCUGAUCCUGCUGGCUUUCGUCCUUUAUUUC ..((((.((((((...((((...............))))..(((.((((....(((((..(((...)))....((....)).)))))....))))..))).)))))).))))........ ( -27.96) >DroYak_CAF1 137310 120 + 1 CUGGACAAAAGCCUCUGGCUUACCAGCCUCGACAUAGACUUGCACAUUAUCCAGGGCCUUGGCUAUGUUCCUUUCUUUCGAUAGAUCCCUCUGAUCCUGUUGGCUUUCGUCCUUUAUUUC ..((((.((((((...((((....))))..(((((((.((.((.(........).))...)))))))))..........((((((((.....))).))))))))))).))))........ ( -30.80) >consensus CUGGACAAAAGCCUCUGGCUUACCAGCCUCCACAUAAACUUGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCAUUUGAUAGCUCCUUCCGCUCCUGCUGGCUUUCGUCCUUUAUUUC ..((((..((((.....)))).(((((..............((((........))))...((((((.((((....)))).))))))............))))).....))))........ (-22.50 = -24.50 + 2.00)

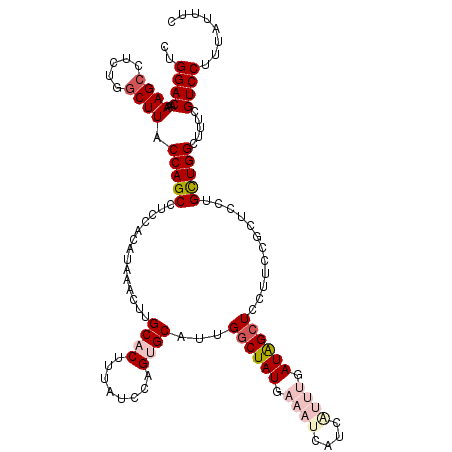

| Location | 9,012,004 – 9,012,124 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -45.34 |
| Consensus MFE | -31.04 |
| Energy contribution | -34.32 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.25 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9012004 120 - 27905053 GAAAUAGAGGACGAAAGCCAGCAGGAGCGGAAUGAGCUAUCAAACGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCAAGUUUAUGUGGAGGCUGGUAAGCCAGAGGCUUUUGUCCAG ........(((((((((((.(((.((((((.(((((.((((....))))..)))))..))..((((((......)))))).)))).)))...((((....))))...))))))))))).. ( -46.30) >DroSec_CAF1 125979 120 - 1 GAAAUAAAGGACGAAAGCCAGCAGGAGCGGAAGGAGCUAUCAAAUGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCAAGUUUAUGUGCAGGCUGGUAAGCCAGACGCUUUUGUCCAG ........((((((((((..(((.((((((..((((.((((....)))).))))....))..((((((......)))))).))))...))).((((....))))....)))))))))).. ( -43.20) >DroSim_CAF1 129219 120 - 1 GAAAUAAAGGACGAAAGCCAGCAGGAGCGGAAGGAGCUAUCAAAUGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCAAGUUUAUGUGCAGGCUGGUAAGCCAGAGGCUUUUGUCCAG ........(((((((((((.(((.((((((..((((.((((....)))).))))....))..((((((......)))))).))))...))).((((....))))...))))))))))).. ( -47.30) >DroEre_CAF1 132018 120 - 1 GAAAUAAAGGACGAAAGCCAGCAGGAUCAGAAGGAGCCAUCGAAGGAAAGGGGCAUAGCCAAGGCUCUGGAUAAUGUGCAAGUCUAUGUGGAGGCUGGUAAGACGGAGGCUUUUGUCCAG ........(((((((((((.(((..(((....((((((.((....))....(((...)))..)))))).)))..)))((.(((((......))))).))........))))))))))).. ( -46.70) >DroYak_CAF1 137310 120 - 1 GAAAUAAAGGACGAAAGCCAACAGGAUCAGAGGGAUCUAUCGAAAGAAAGGAACAUAGCCAAGGCCCUGGAUAAUGUGCAAGUCUAUGUCGAGGCUGGUAAGCCAGAGGCUUUUGUCCAG ........(((((((((((.(((..(((..((((.(((.((....))..((.......)).))))))).)))..)))...............((((....))))...))))))))))).. ( -43.20) >consensus GAAAUAAAGGACGAAAGCCAGCAGGAGCGGAAGGAGCUAUCAAAUGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCAAGUUUAUGUGGAGGCUGGUAAGCCAGAGGCUUUUGUCCAG ........(((((((((((.(((.((((....((((.((((....)))).)))).........(((((......)))))..)))).)))...((((....))))...))))))))))).. (-31.04 = -34.32 + 3.28)


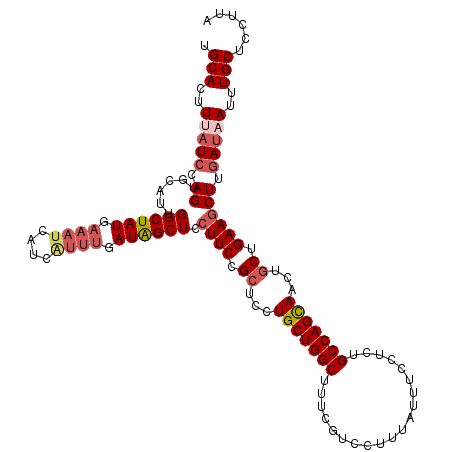
| Location | 9,012,044 – 9,012,164 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -26.23 |
| Energy contribution | -28.23 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9012044 120 + 27905053 UGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCGUUUGAUAGCUCAUUCCGCUCCUGCUGGCUUUCGUCCUCUAUUUCCUGUGCCAGCAACUGCUGAAGCUUGAUCAUUUGCUCCUUG ((((((......))))))..((((((.((((....)))).))))))..(((.((...(((((((....................)))))))...)).)))((..........))...... ( -30.95) >DroSec_CAF1 126019 120 + 1 UGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCAUUUGAUAGCUCCUUCCGCUCCUGCUGGCUUUCGUCCUUUAUUUCCUCUGCCAGCAACUGCUGAAGCUUGAUAAUUUGCUCCUUA .(((..(((((.((......((((((.((((....)))).)))))).((((.((...(((((((....................)))))))...)).)))))).)))))..)))...... ( -34.05) >DroSim_CAF1 129259 120 + 1 UGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCAUUUGAUAGCUCCUUCCGCUCCUGCUGGCUUUCGUCCUUUAUUUCCUCUGCCAGCAACUGCUGAAGCUUGAUAAUUUGCUCCUUA .(((..(((((.((......((((((.((((....)))).)))))).((((.((...(((((((....................)))))))...)).)))))).)))))..)))...... ( -34.05) >DroEre_CAF1 132058 120 + 1 UGCACAUUAUCCAGAGCCUUGGCUAUGCCCCUUUCCUUCGAUGGCUCCUUCUGAUCCUGCUGGCUUUCGUCCUUUAUUUCCUCUGCCAGUAACUGCUGAAGCUUUAGCAUCUGCUCCUUA .............((((....((((.(((....((....)).)))..((((.(....(((((((....................)))))))....).))))...))))....)))).... ( -25.45) >consensus UGCACUUUAUCCAGUGCAUUGGCUAUGAAAUCAUCAUUUGAUAGCUCCUUCCGCUCCUGCUGGCUUUCGUCCUUUAUUUCCUCUGCCAGCAACUGCUGAAGCUUGAUAAUUUGCUCCUUA .(((..(((((.((......((((((.((((....)))).)))))).((((.((...(((((((....................)))))))...)).)))))).)))))..)))...... (-26.23 = -28.23 + 2.00)


| Location | 9,012,044 – 9,012,164 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -24.45 |
| Energy contribution | -25.88 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9012044 120 - 27905053 CAAGGAGCAAAUGAUCAAGCUUCAGCAGUUGCUGGCACAGGAAAUAGAGGACGAAAGCCAGCAGGAGCGGAAUGAGCUAUCAAACGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCA ...(((((..........))))).((..((((((((....................))))))))..))((.(((((.((((....))))..)))))..))..((((((......)))))) ( -33.95) >DroSec_CAF1 126019 120 - 1 UAAGGAGCAAAUUAUCAAGCUUCAGCAGUUGCUGGCAGAGGAAAUAAAGGACGAAAGCCAGCAGGAGCGGAAGGAGCUAUCAAAUGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCA ...((...(((((((((((((((.((..((((((((....................))))))))..))....))))))......))))))))).....))..((((((......)))))) ( -36.95) >DroSim_CAF1 129259 120 - 1 UAAGGAGCAAAUUAUCAAGCUUCAGCAGUUGCUGGCAGAGGAAAUAAAGGACGAAAGCCAGCAGGAGCGGAAGGAGCUAUCAAAUGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCA ...((...(((((((((((((((.((..((((((((....................))))))))..))....))))))......))))))))).....))..((((((......)))))) ( -36.95) >DroEre_CAF1 132058 120 - 1 UAAGGAGCAGAUGCUAAAGCUUCAGCAGUUACUGGCAGAGGAAAUAAAGGACGAAAGCCAGCAGGAUCAGAAGGAGCCAUCGAAGGAAAGGGGCAUAGCCAAGGCUCUGGAUAAUGUGCA ......(((.(((((....((((.((........)).)))).......((.(....))))))...(((....((((((.((....))....(((...)))..)))))).))).)).))). ( -33.90) >consensus UAAGGAGCAAAUGAUCAAGCUUCAGCAGUUGCUGGCAGAGGAAAUAAAGGACGAAAGCCAGCAGGAGCGGAAGGAGCUAUCAAAUGAUGAUUUCAUAGCCAAUGCACUGGAUAAAGUGCA ...((........((((((((((.((..((((((((....................))))))))..))....)))))).((....)))))).......))...(((((......))))). (-24.45 = -25.88 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:56 2006