| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,952,129 – 8,952,324 |
| Length | 195 |
| Max. P | 0.594006 |

| Location | 8,952,129 – 8,952,219 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.27 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8952129 90 + 27905053 CUUGUCCGCCAGCGAGGAGCUAUAGAAUUGCUGGCAGCGUUGCUGU-----UGCAGUUCCCGCGCUUGUAGAUUGUGCAGCGGGGAGCUGUAGGG .....((((((((((....(....)..))))))))...........-----((((((((((.((((.(((.....))))))))))))))))))). ( -39.70) >DroVir_CAF1 92388 91 + 1 --UGUCCGCCAGGGAGGCACUGUAGAACUGUAGCUUCUGCUGCU--CGCGCUGCAGUUCACGUGCCUGCAAAUUGUGCAGCGGCGAGCUGUAUGG --......((..(.((((((.((.((((((((((....((....--.)))))))))))))))))))).).....(((((((.....))))))))) ( -39.30) >DroSec_CAF1 74145 90 + 1 CUUGUCCGCCAGCGAGGAGCUAUAGAACUGCUGGCAGCGUUGCUGU-----UGCAGUUCCCGCGCUUGCAGAUUGUGCAGCGGGGAGCUGUAGGG .....((((((((.((...(....)..))))))))...........-----((((((((((.((((.(((.....))))))))))))))))))). ( -40.80) >DroEre_CAF1 79520 90 + 1 UCUGUCCGCCAGAGAGGAGCUGUAGAAUUGCUGGCAGCGUUGCUGU-----UGCAGUUCGCGCGCUUGUAGAUUGUGCAGCGGGGAGCUGUAGGG ((((.....)))).(.(((((((.(((((((.((((((...)))))-----))))))))))).)))).)....(.((((((.....)))))).). ( -31.80) >DroYak_CAF1 80515 90 + 1 CUUGUCCGCCAGCGAGGAGCUGUAGAAUUGCUGGCAGCGCUGCUGU-----UGCAGUUCGCGCGCUUGUAGAUUGUGCAGCGGGGAGCUGUAGGG .....((..((((.....((((((.((((((.(((.(((((((...-----.))))..)))..))).))).))).)))))).....))))..)). ( -36.70) >DroMoj_CAF1 98604 91 + 1 --UGUCCGCUAGAGAAGCGCUGUAGAACUGCAGCUUCUGUUGCU--CGCGCUGCAGCUCACGUGCCUGCAGAUUGUGCAGCGGGGAGCUGUAGGG --....((((.....))))........((((((((((((((((.--((..(((((((......).))))))..)).))))).))))))))))).. ( -40.00) >consensus CUUGUCCGCCAGCGAGGAGCUGUAGAACUGCUGGCAGCGUUGCUGU_____UGCAGUUCACGCGCUUGCAGAUUGUGCAGCGGGGAGCUGUAGGG .....(((((((...((..(....)..)).)))))................((((((((.(.(((.((((.....))))))).))))))))))). (-23.76 = -23.27 + -0.50)

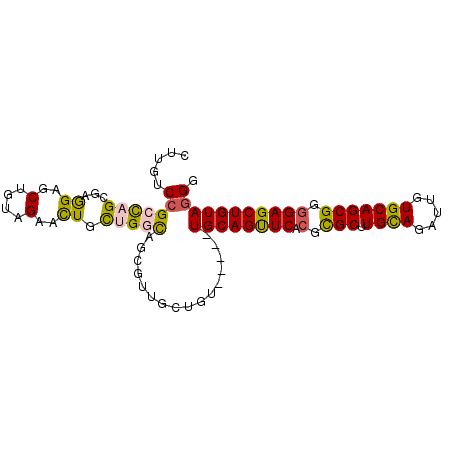
| Location | 8,952,219 – 8,952,324 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -24.62 |
| Energy contribution | -23.23 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8952219 105 + 27905053 UGCUUCUCUCCGCGUGCUGUGGUGCCACUUUUGCAGGCGAGCCGGAUCACCGCCAUUGGAGUUCACAGAUCCCUGAUAGUGCAGGAAGCUGUGCGCAUCAUCGGU ..........(((.(((.(((....)))....))).))).(((((....(((....))).((.(((((...((((......))))...))))).))....))))) ( -34.10) >DroPse_CAF1 79092 102 + 1 CGCCUCGCGGCGGGUGCUGUGGUGCCAUUUCUGGAGCCGUGCAGGAUCUGCUG---UGCUGCCUGCAGAUCCCUGAUAGUUCAGGAAGCUGUGGGCGUCGUCGCU .((..((((((((((.(((((((.(((....))).)))..)))).))))))))---))..(((..(((...(((((....)))))...)))..)))......)). ( -49.80) >DroWil_CAF1 77541 105 + 1 AGCCUCUCGUCUGGUACUGUGAUGCCAUUUCUGUAGUCGGGCCGGAUCUGCUGGAUGAUUGCCACUAGAUCCUUGAUAAUGCAAAAAACUAUGUCCAUCAUCACU .(((........)))...((((((.((....)).....((((.(((((((.(((.......))).)))))))(((......)))........))))..)))))). ( -25.10) >DroYak_CAF1 80605 105 + 1 AGCUUCUCGCCGCGUGCUGUGGUGCCACUUUUGCAGGCGAGCCGGAUCUCCCCCAUUGGAGUUCACAGAUCCCUGAUAGUGCAGGAAGCUGUGCGCAUCAUCCGU .....(((((((((....(((....)))...))).)))))).(((((.(((......)))((.(((((...((((......))))...))))).))...))))). ( -38.30) >DroAna_CAF1 79514 105 + 1 CGCCUCUCUUCGAGUGCUGUGAUGCCACUUUUGCAGGCGGGCCGGAUCCGUCGAAUUAGAGUUGACGGAUCCCUGAUAGUGCAGGAAGCUGUGCACAUCAUCACU .(((((.....))).)).((((((((.((......)).))...(((((((((((.......)))))))))))......(..(((....)))..)....)))))). ( -39.80) >DroPer_CAF1 80344 102 + 1 CGCCUCGCGGCGGGUGCUGUGGUGCCAUUUCUGGAGCCGUGCAGGAUCUGCUG---UGCUGCCUGCAGAUCCCUGAUAGUUCAGGAAGCUGUGGGCGUCGUCGCU .((..((((((((((.(((((((.(((....))).)))..)))).))))))))---))..(((..(((...(((((....)))))...)))..)))......)). ( -49.80) >consensus CGCCUCUCGCCGGGUGCUGUGGUGCCACUUCUGCAGGCGAGCCGGAUCUGCUG_AUUGGAGCCCACAGAUCCCUGAUAGUGCAGGAAGCUGUGCGCAUCAUCGCU .....((((((.......(((....))).......))))))..(((((((...............))))))).((((.(((((.(....).)))))))))..... (-24.62 = -23.23 + -1.38)

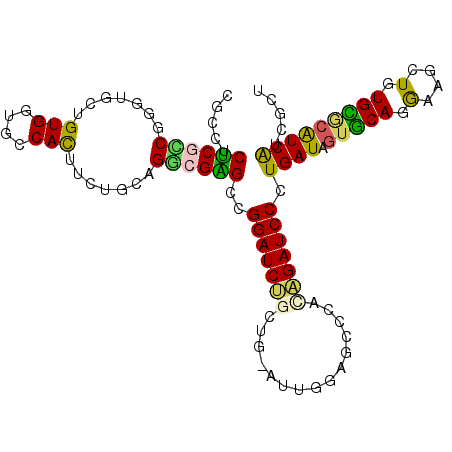

| Location | 8,952,219 – 8,952,324 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.38 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8952219 105 - 27905053 ACCGAUGAUGCGCACAGCUUCCUGCACUAUCAGGGAUCUGUGAACUCCAAUGGCGGUGAUCCGGCUCGCCUGCAAAAGUGGCACCACAGCACGCGGAGAGAAGCA ........(((.(((((.((((((......)))))).)))))..((((...(.(((....))).)..((.(((....(((....))).))).))))))....))) ( -36.50) >DroPse_CAF1 79092 102 - 1 AGCGACGACGCCCACAGCUUCCUGAACUAUCAGGGAUCUGCAGGCAGCA---CAGCAGAUCCUGCACGGCUCCAGAAAUGGCACCACAGCACCCGCCGCGAGGCG .........(((..(((.(((((((....))))))).)))..))).((.---..((((...))))..((..(((....)))..))...))...((((....)))) ( -40.30) >DroWil_CAF1 77541 105 - 1 AGUGAUGAUGGACAUAGUUUUUUGCAUUAUCAAGGAUCUAGUGGCAAUCAUCCAGCAGAUCCGGCCCGACUACAGAAAUGGCAUCACAGUACCAGACGAGAGGCU ..((((((((.(.(......).).)))))))).((((((..(((.......)))..))))))(((((((...((....))...))...((.....))..).)))) ( -24.60) >DroYak_CAF1 80605 105 - 1 ACGGAUGAUGCGCACAGCUUCCUGCACUAUCAGGGAUCUGUGAACUCCAAUGGGGGAGAUCCGGCUCGCCUGCAAAAGUGGCACCACAGCACGCGGCGAGAAGCU .(((((......(((((.((((((......)))))).)))))..((((......))))))))).((((((.((....(((....))).....))))))))..... ( -42.10) >DroAna_CAF1 79514 105 - 1 AGUGAUGAUGUGCACAGCUUCCUGCACUAUCAGGGAUCCGUCAACUCUAAUUCGACGGAUCCGGCCCGCCUGCAAAAGUGGCAUCACAGCACUCGAAGAGAGGCG .(((((((((((((........))))).)))..(((((((((...........))))))))).....(((.((....)))))))))).((.(((.....))))). ( -39.80) >DroPer_CAF1 80344 102 - 1 AGCGACGACGCCCACAGCUUCCUGAACUAUCAGGGAUCUGCAGGCAGCA---CAGCAGAUCCUGCACGGCUCCAGAAAUGGCACCACAGCACCCGCCGCGAGGCG .........(((..(((.(((((((....))))))).)))..))).((.---..((((...))))..((..(((....)))..))...))...((((....)))) ( -40.30) >consensus AGCGAUGAUGCGCACAGCUUCCUGCACUAUCAGGGAUCUGUAAACACCAAU_CAGCAGAUCCGGCACGCCUGCAAAAAUGGCACCACAGCACCCGACGAGAGGCG ................((((((((......)))((((((.................))))))....((((..((....))((......))....)))).))))). (-17.90 = -17.38 + -0.52)

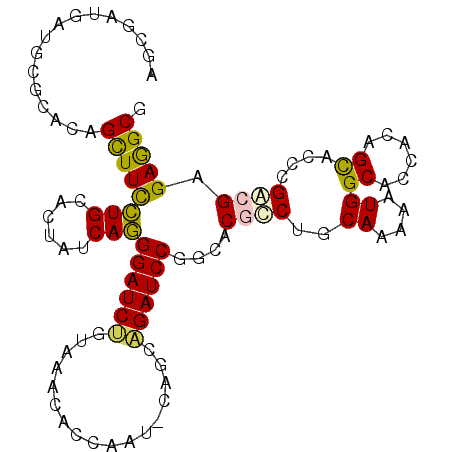
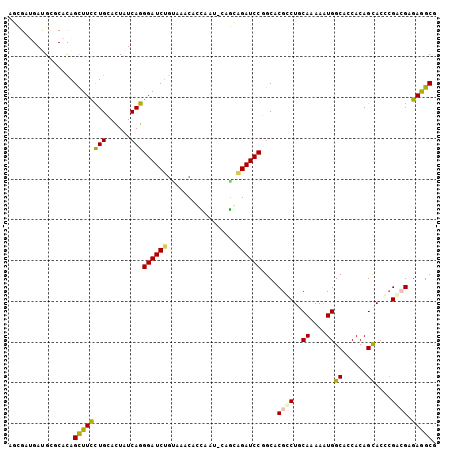
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:36 2006