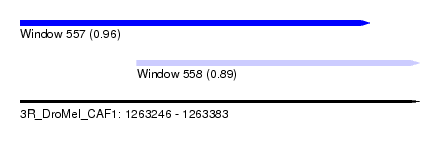
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,263,246 – 1,263,383 |
| Length | 137 |
| Max. P | 0.963843 |

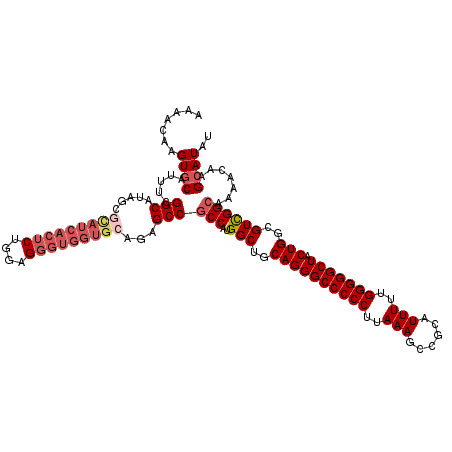
| Location | 1,263,246 – 1,263,366 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -47.97 |
| Consensus MFE | -40.74 |
| Energy contribution | -42.27 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1263246 120 + 27905053 AGAACAAGUGCAUCUCGGCAUAGCGCAUCACUCUGGAGGGUGGUGCGGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCGUUUUUGGGGGCUACUGGGGUCGGCAAAACAAGCAUAC .......((((.....(((....((((((((((....))))))))))..)))(((..((((.((((((((((..(((((...))))).))))))).))).))))))).......)))).. ( -56.00) >DroVir_CAF1 19501 120 + 1 AAAAUAAGUGCAUUUCGGCAUAGCGUAUCACUCUGGAGGGUGGUGGAGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCAGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAU .......((((.....(((.(..(.((((((((....)))))))))..))))(((..(((.((..(((((((..(((......)))..)))))))....)))))))).......)))).. ( -44.50) >DroGri_CAF1 33744 120 + 1 AAAAUAAGUGCAUUUCGGCAUAGCGUAUCACUCUGGAGGGUGGUGCAGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCUGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAU .......((((.....(((.....(((((((((....)))))))))...)))(((..(((.((..(((((((..(((......)))..)))))))....)))))))).......)))).. ( -46.80) >DroSec_CAF1 40095 120 + 1 AGAACAAGUUCAUCUCGGCAUAGCGCAUCACUCUGGAGGGUGGUGCGGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCGUUUUUGGGGGCUACUGGGGUCGGCAAAACAAGCAUAC .(((....))).....(((....((((((((((....))))))))))..)))(((..((((.((((((((((..(((((...))))).))))))).))).)))))))............. ( -52.40) >DroWil_CAF1 26116 111 + 1 AAAAUAAGUGCAUUUCGGCAUAGCGCAUCACUCUGGAG---------GAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCAUUUUUGGGGGCUACUGGCGUUGGCAAAACAAGCAUAU .......((((......((.((((((............---------.((((.....)))).((((((((((..(((......)))..))))))).))))))))))).......)))).. ( -38.12) >DroMoj_CAF1 29261 120 + 1 AAAACAAGUGCAUUUCGGCAUAGCGUAUCACUCUGGAGGGUGGUGCAGAGCCCCCAUGGCUGCAGGGCCCCCUUAAAGCAGCAUUUUUGGGGGCUUCUGGCGUCGGCAAAACAAGCAUAU .......((((......((...(((((((((((....))))))))).(((((((((..((((((((....)))....))))).....)))))))))...))....)).......)))).. ( -50.02) >consensus AAAACAAGUGCAUUUCGGCAUAGCGCAUCACUCUGGAGGGUGGUGCAGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAU .......((((.....(((.....(((((((((....)))))))))...)))(((..(((..((((((((((..(((......)))..))))))).)))..)))))).......)))).. (-40.74 = -42.27 + 1.53)

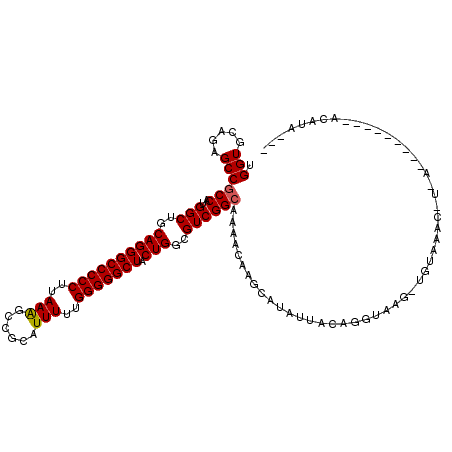
| Location | 1,263,286 – 1,263,383 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1263286 97 + 27905053 UGGUGCGGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCGUUUUUGGGGGCUACUGGGGUCGGCAAAACAAGCAUACUACAGGUGAG-UCCAAAC--------------------- ......(((..((((.(((((.((((((((((..(((((...))))).))))))).))).)))))((.......))........))))..-)))....--------------------- ( -38.70) >DroVir_CAF1 19541 107 + 1 UGGUGGAGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCAGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAUUACAGGUAAG-UGUUAUCUUCA--------CAUAUA--- ..((((((((((((....)).((..(((((((..(((......)))..)))))))....))...)))......(((((.(((....))))-)))).))))))--------).....--- ( -39.00) >DroGri_CAF1 33784 110 + 1 UGGUGCAGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCUGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAUUACAAGUAUG-UAUUUACUGCA--------AACAUAUGU ...(((((.(((((....)).((..(((((((..(((......)))..)))))))....))...))).......((((((.....)))))-).....)))))--------......... ( -38.80) >DroYak_CAF1 52694 117 + 1 UGGUGCGGAGCCGCCAUGGCUGCAGGGCCCCCUUAAGGCCGCGUUUUUGGGGGCUACUGGGGUCGGCAAAACAAGCAUACUACAGGUGAG-UCCAAAU-UUACCAUGGUUCACCUAUGU ..(((((....)(((..((((.((((((((((..(((((...))))).))))))).))).))))))).......)))).....(((((((-.(((...-......)))))))))).... ( -45.70) >DroMoj_CAF1 29301 103 + 1 UGGUGCAGAGCCCCCAUGGCUGCAGGGCCCCCUUAAAGCAGCAUUUUUGGGGGCUUCUGGCGUCGGCAAAACAAGCAUAUUACAGGUAAACUGA-----UAA--------UACGUA--- .(..((((((((((((..((((((((....)))....))))).....))))))).))).))..)..........((.((((((((.....))).-----)))--------)).)).--- ( -40.20) >DroAna_CAF1 39175 106 + 1 UGGUGCGGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAUUACAGGUAAG-CGUAAAA-U-----------UAUUUUUU ((.(((.(((((((....)).....(((((((..(((......)))..)))))))...))).)).)))...)).((.(((.....))).)-)......-.-----------........ ( -33.20) >consensus UGGUGCAGAGCCGCCAUGGCUGCAGGGCCCCCUUAAAGCCGCAUUUUUGGGGGCUACUGGCGUCGGCAAAACAAGCAUAUUACAGGUAAG_UGUAAAC_U_A_________ACAUA___ .(((.....)))(((..(((..((((((((((..(((......)))..))))))).)))..)))))).................................................... (-28.00 = -28.03 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:39 2006