
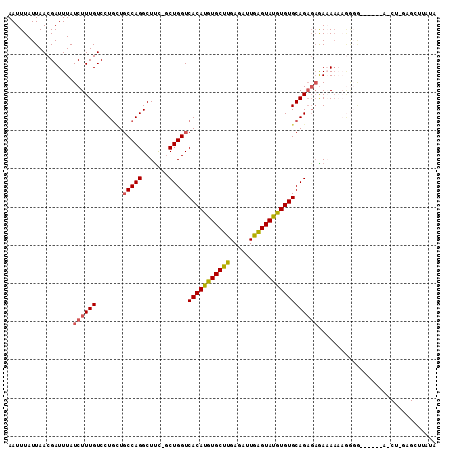
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,929,130 – 8,929,260 |
| Length | 130 |
| Max. P | 0.989958 |

| Location | 8,929,130 – 8,929,241 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8929130 111 + 27905053 AAUUUAUUAACGAUUUAUCUUUGUCCUGCUGCCAGGCUUU-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGCGAAAAAAGGGGGUGUUGAGCUAGAGCUUAUA ...................((((..((((.(((((.....-.))))).((((((((((......))))))))))))))..))))............(((((....))))).. ( -29.70) >DroSec_CAF1 50752 93 + 1 AAUUUAUUAACGAUUUAUCUUUGUCCUGCUGCCAGGCUUC--CUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAAACAAAAGGGG-----------------AUA .................(((((.((((((.((((((...)--))))).((((((((((......)))))))))))))).))....)))))..-----------------... ( -26.50) >DroSim_CAF1 52078 94 + 1 AAUUUAUUAACGAUUUAUCUUUGUCCUUCUGCCAGGCUUC-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAAACAAAAGGGG-----------------AUA ............((((...(((((..((((((..((((..-...))))((((((((((......))))))))))))))))..)))))...))-----------------)). ( -24.60) >DroEre_CAF1 57198 108 + 1 AAUUUAUUAACGAUUUAUCUUUGUCCUGUUGCCAGGCUUC-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGACAGAAAAAAGGUGGUAUUAAACUGGAGCUU--- ...........(((((((((((...((((((((((.....-.)))))(((((((((((......)))))))))))..)))))...)))))))).)))............--- ( -28.10) >DroYak_CAF1 57395 95 + 1 AAUUUAUUAACGAUUUAUCUUUGUCCUGCUGCCAGGCUUC-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAGAAA-------------AACUAGAGCUU--- ...................(((.((((((.(((((.....-.))))).((((((((((......)))))))))))))).)).)))-------------...........--- ( -24.00) >DroAna_CAF1 56303 108 + 1 AAUUUAUUAACGAUUUAUCUUUGUCCUGGUCCCAGGCUCCUUCUGGUCACAUAUGCUUGAGAUUAGGUAUAUGUGCAGUGAGAAAAUUAAGA----AGAGCUGUAGUUUCAA ........(((......(((((..((((....))))(((...(((..((((((((((((....))))))))))))))).)))........))----)))......))).... ( -26.60) >consensus AAUUUAUUAACGAUUUAUCUUUGUCCUGCUGCCAGGCUUC_GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAGAAAAAAGGGG______A_CU_GAGCUUAUA .................((((((.......(((((.......)))))(((((((((((......)))))))))))))))))............................... (-19.54 = -19.65 + 0.11)


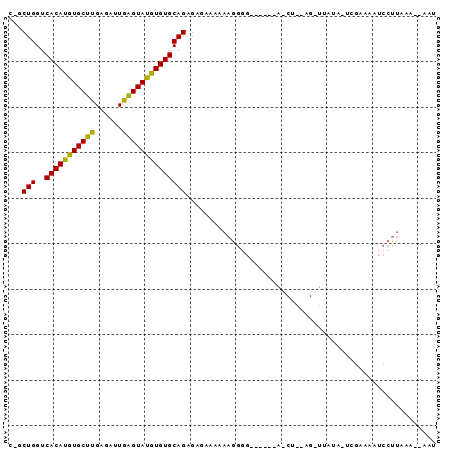
| Location | 8,929,169 – 8,929,260 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 66.74 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -14.24 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8929169 91 + 27905053 U-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGCGAAAAAAGGGGGUGUUGAGCUAGAGCUUAUAUUCGAAAAUCCUUAAA--AAU .-(((.(.(((((((((((......))))))))))))..))).....(((((..((.(((((....)))))....))....)))))...--... ( -25.10) >DroSec_CAF1 50791 73 + 1 C--CUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAAACAAAAGGGG-----------------AUAGUCGAAAAUCCUUAUA--GAU .--(((..(((((((((((......))))))))))))))..........((((-----------------((........))))))...--... ( -18.70) >DroSim_CAF1 52117 74 + 1 C-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAAACAAAAGGGG-----------------AUAGUCGAAAAUCCUUAAA--GAU .-.(((..(((((((((((......))))))))))))))..........((((-----------------((........))))))...--... ( -18.60) >DroEre_CAF1 57237 83 + 1 C-GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGACAGAAAAAAGGUGGUAUUAAACUGGAGCUU----------AUCCUUACAUUAAU .-.(((..(((((((((((......)))))))))))....)))....((((.(((...........))).----------..))))........ ( -18.00) >DroAna_CAF1 56342 77 + 1 CUUCUGGUCACAUAUGCUUGAGAUUAGGUAUAUGUGCAGUGAGAAAAUUAAGA----AGAGCUGUAGUUUCAA-----------UUAUA--UAU (((((.(.((((((((((((....)))))))))))))(((......))).)))----))...((((((....)-----------)))))--... ( -16.70) >consensus C_GCUGGUCACAUGUGCUUGAGAUUGAGUAUGUGUGCAGAGAGAAAAAAGGGG______A_CU__AG_UUAUA_UCGAAAAUCCUUAAA__AAU ...(((..(((((((((((......))))))))))))))....................................................... (-14.24 = -13.60 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:28 2006