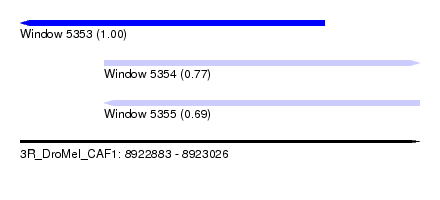
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,922,883 – 8,923,026 |
| Length | 143 |
| Max. P | 0.996997 |

| Location | 8,922,883 – 8,922,992 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8922883 109 - 27905053 GAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGC---CAGCGAAA--GA-GACC---- (((((((((((.....((..-.)).....))))))))))).((((....(((((((.............)))))))....))))............---...(....--).-....---- ( -19.72) >DroVir_CAF1 56845 113 - 1 GAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACAGAGCA--CAACGAGC--AAUGACCAA-- (((((((((((.....((..-.)).....))))))))))).((((....(((((((.............)))))))....))))......((..((.--......))--..)).....-- ( -20.72) >DroGri_CAF1 54363 118 - 1 GAAUGACAACAUUUUUGCAAAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACUCAGCA--CAACGAGCACAAUGACCACGA (((((((((((.....((....)).....))))))))))).((((....(((((((.............)))))))....))))......(((....--....))).............. ( -21.72) >DroWil_CAF1 49884 108 - 1 GAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGAC--CA--GAAA--GG-GAGU---- (((((((((((.....((..-.)).....))))))))))).........(((((((.............))))))).....(((((...........--..--....--))-))).---- ( -21.15) >DroMoj_CAF1 58916 113 - 1 GAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUUAAGAAAUUUCCCACCACACAGCG--CAACGAGC--AUUGGCCAA-- (((((((((((.....((..-.)).....))))))))))).((((....(((((((((..........)))))))))...))))........(((.(--(.....))--.))).....-- ( -21.40) >DroPer_CAF1 50249 113 - 1 GAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCGGACGGCGGAC--GACGAGC---- (((((((((((.....((..-.)).....))))))))))).((((....(((((((.............)))))))....))))((.((...........)).))..--.......---- ( -22.12) >consensus GAAUGACAACAUUUUUGCAA_AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCA__CAACGAAC__AAUGACC____ (((((((((((.....((....)).....))))))))))).((((....(((((((.............)))))))....)))).................................... (-19.94 = -20.10 + 0.17)

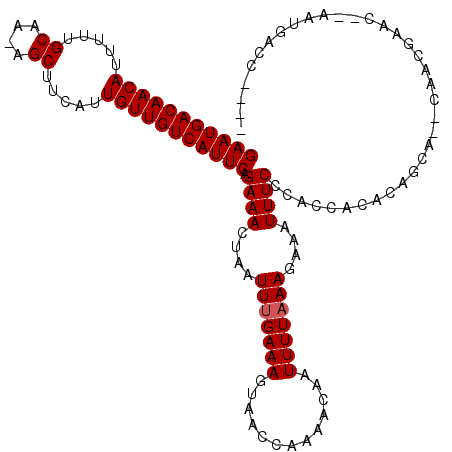
| Location | 8,922,913 – 8,923,026 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8922913 113 + 27905053 UUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCU-UUGCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAGAU------CCUACCCUUU ...............(((((..(((((.......(((.(.(((((((((((.....((.-..)).....))))))))))).)..)))..)))))....))))).------.......... ( -19.70) >DroVir_CAF1 56879 119 + 1 UUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCU-UUGCAAAAAUGUUGUCAUUCCGUUGCUGCAAAGUUUUCACAACCGGGCACCCUACCCCAA .............(((.(((..(((((.....((((..(.(((((((((((.....((.-..)).....))))))))))).)..)))).)))))..))).))).(((........))).. ( -27.20) >DroGri_CAF1 54401 120 + 1 UUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUUUGCAAAAAUGUUGUCAUUCCGCUGCUGUAAAGUUUUCUCAGUCAGACGCCCAACAACGU .............(((((((....................(((((((((((.....((....)).....)))))))))))((((((((...........))).))).)).)))).))).. ( -25.90) >DroWil_CAF1 49913 101 + 1 UUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCU-UUGCAAAAAUGUUGUCAUUCCGUUGGAGUAAAGUUUUGCCAU------------------ .................((((.(((((...(((.....(.(((((((((((.....((.-..)).....))))))))))).)....))))))))...)))).------------------ ( -20.10) >DroMoj_CAF1 58950 107 + 1 UUUCUUAAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCU-UUGCAAAAAUGUUGUCAUUCCGUUGCUGCAAACUUUUCUCAACCAGCC------------ ................((((((..........((((..(.(((((((((((.....((.-..)).....))))))))))).)..))))............))))))..------------ ( -21.35) >DroAna_CAF1 50903 108 + 1 UUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCU-UUGCAAAAAUGUUGUCAUUCCGUUGGCGCAAACUUUUGCCUU-----------GCUCCUC ........((((((.(((((......))))).))))))..(((((((((((.....((.-..)).....))))))))))).((.((.((((....)))))).-----------))..... ( -22.80) >consensus UUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCU_UUGCAAAAAUGUUGUCAUUCCGUUGCUGCAAAGUUUUCCCAA_C_________AC_CC__ ........((((((.(((((......))))).))))))..(((((((((((.....((....)).....)))))))))))........................................ (-19.90 = -19.90 + 0.00)

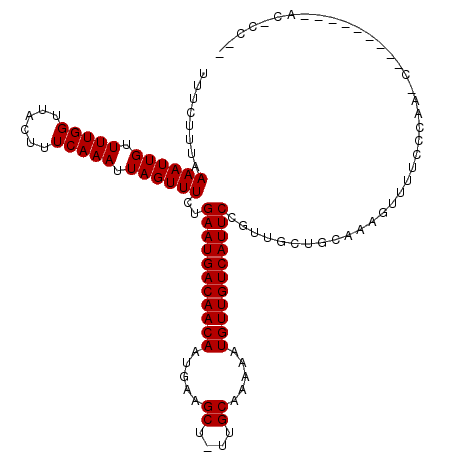
| Location | 8,922,913 – 8,923,026 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
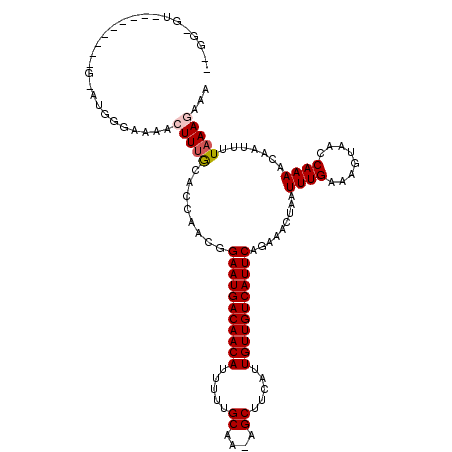
>3R_DroMel_CAF1 8922913 113 - 27905053 AAAGGGUAGG------AUCUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAA ...((((.((------....((.....)).....)).)).(((((((((((.....((..-.)).....)))))))))))....................)).................. ( -21.60) >DroVir_CAF1 56879 119 - 1 UUGGGGUAGGGUGCCCGGUUGUGAAAACUUUGCAGCAACGGAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAA ...((((.....)))).((((..(.....)..))))....(((((((((((.....((..-.)).....))))))))))).........(((((((.............))))))).... ( -28.02) >DroGri_CAF1 54401 120 - 1 ACGUUGUUGGGCGUCUGACUGAGAAAACUUUACAGCAGCGGAAUGACAACAUUUUUGCAAAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAA ..(((.((((.((.(((.((((((....))).))))))))(((((((((((.....((....)).....)))))))))))....................)))))))............. ( -30.20) >DroWil_CAF1 49913 101 - 1 ------------------AUGGCAAAACUUUACUCCAACGGAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAA ------------------.(((....(((((.........(((((((((((.....((..-.)).....))))))))))).............)))))..)))................. ( -18.85) >DroMoj_CAF1 58950 107 - 1 ------------GGCUGGUUGAGAAAAGUUUGCAGCAACGGAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUUAAGAAA ------------...((((((....(((((..........(((((((((((.....((..-.)).....))))))))))).......))))).....))))))................. ( -22.43) >DroAna_CAF1 50903 108 - 1 GAGGAGC-----------AAGGCAAAAGUUUGCGCCAACGGAAUGACAACAUUUUUGCAA-AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAA ..((.((-----------(((.(....)))))).))....(((((((((((.....((..-.)).....))))))))))).........(((((((.............))))))).... ( -23.72) >consensus __GG_GU_________G_AUGGGAAAACUUUGCACCAACGGAAUGACAACAUUUUUGCAA_AGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAA ...........................(((((........(((((((((((.....((....)).....))))))))))).........((((........)))).......)))))... (-18.20 = -18.48 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:22 2006