| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,864,945 – 8,865,049 |
| Length | 104 |
| Max. P | 0.713363 |

| Location | 8,864,945 – 8,865,049 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.59 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.50 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8864945 104 + 27905053 CUGCGCCAUGAUUGCGCCUCGGUUCGCCAUAACGGCAGCACACUGUGCCAGCGUCGGUGGGUAAGCAAAAAAAAAUAAAUAAAAAUGAAAAACAAAACCAAAGA----- ..((((.......))))...((((.(((.....))).((.(((((((....)).))))).)).................................)))).....----- ( -20.50) >DroSec_CAF1 397 89 + 1 CUGCGCCAUGAUUGCGCCUCGGUUCGCCAUAACGGCAGCCCACUGUGCCAGCGUCGGUGGGUAAGCAAAAAAAAGA-------A---GAAAACAA-----AACA----- .((((((...((.(((........))).))...))).((((((((((....)).))))))))..))).........-------.---........-----....----- ( -25.30) >DroSim_CAF1 389 92 + 1 CUGCGCCAUGAUUGCGCCUCGGUUCGCCAUAACGGCAGCCCACUGUGCCAGCGUCGGUGGGUAAGCAAAAAAAAAA-------AAAGAAAAACAA-----CACA----- .((((((...((.(((........))).))...))).((((((((((....)).))))))))..))).........-------............-----....----- ( -25.30) >DroEre_CAF1 474 96 + 1 CUGCGUUAUGAUUGCGCCUCGGUUCGCCAUAACGGCAGCCCACUGUGCCAGCGUCGGUGGGUAAGCAAAAAAUG-A-------AAAGAAAAAUAC-----GAUAGAGCG ..((((.......))))....(((((((.....))).((((((((((....)).))))))))............-.-------............-----....)))). ( -27.40) >DroYak_CAF1 404 96 + 1 CUGCGUCAUGAUUGCGCCUCGGUUCGCCAUAACGGCAGCCCACUGUGCCAGCGUCGGUGGGUAAGCAAAAAACG-A-------AAAGAAAAACAC-----GAUGGAGCG .............((.(((((.((((((.....))).((((((((((....)).))))))))............-.-------...))).....)-----)).)).)). ( -28.60) >DroAna_CAF1 474 100 + 1 UUGCGUAUUAAUUGCCCCAAAGUUUGCUUUAACGGCCGCCCACUGUGCCAACAUUGGCGGGUAAGUGACAAAGAGA---GAAAUAUAACGAACAU-ACUAAAAU----- ....((.....((((((.((((....)))).((((.......))))((((....))))))))))...)).......---................-........----- ( -16.40) >consensus CUGCGCCAUGAUUGCGCCUCGGUUCGCCAUAACGGCAGCCCACUGUGCCAGCGUCGGUGGGUAAGCAAAAAAAA_A_______AAAGAAAAACAA_____AACA_____ ...........((((..........(((.....))).((((((((((....)).))))))))..))))......................................... (-19.86 = -19.50 + -0.36)

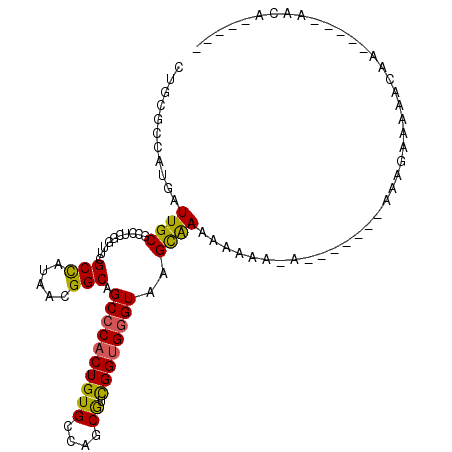

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:03 2006