
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
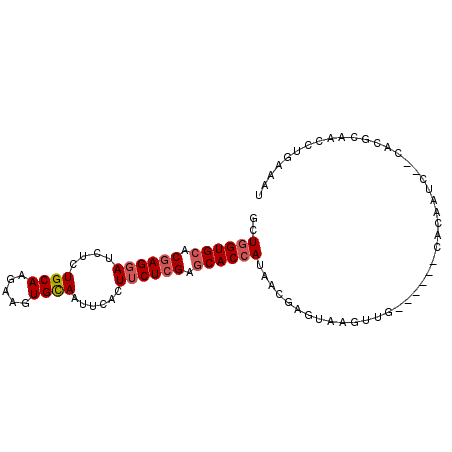
| Location | 8,826,638 – 8,826,733 |
| Length | 95 |
| Max. P | 0.737137 |

| Location | 8,826,638 – 8,826,733 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8826638 95 + 27905053 GCUGGUGCACGAGGAUCUCUGCAAGAAGUGCAAUUCACUUCUCGAGCACCAUAACGAGUAAGUAGAUUGGGUAGAUUGUGCACCCAAUCUGAAAU ..((((((.((((((....((((.....))))......)))))).))))))...........((((((((((.(......))))))))))).... ( -33.10) >DroVir_CAF1 1531 85 + 1 GCUGGUGCACGAGGACCUAUGCAAGAAGUGUAAUGCAAUGCUGGAACACCAUAAUGAGUGAGUGA------UUCA-UC--GAUAUAAC-UGCAGC ((((((.(....).)))...(((...(((((......))))).(((((((((.....))).))).------))).-..--........-)))))) ( -14.90) >DroSec_CAF1 1105 87 + 1 GCUGGUGCACGAGGAUCUCUGCAAGAAGUGCAAUUCACUUCUCGAGCACCAUAACGAGUAAGUUG------CACAAUC--UACGCAACCUGAAAU ..((((((.((((((....((((.....))))......)))))).))))))..........((((------(......--...)))))....... ( -24.70) >DroSim_CAF1 1062 87 + 1 GCUGGUGCACGAGGAUCUCUGCAAGAAGUGCAAUUCACUUCUCGAGCACCAUAACGAGUAAGUUG------CACAAUC--UGCGCAACCUGAAAU ..((((((.((((((....((((.....))))......)))))).))))))..........((((------(.(....--.).)))))....... ( -26.20) >DroEre_CAF1 1130 87 + 1 GCUGGUGCACGAGGAUCUCUGCAAGAAAUGCAAUUCUCUUCUCGAGCACCAUAACGAGUAGGUUU------CACGAGC--CUCGCGACCUGAAAU ..((((((.((((((....((((.....))))......)))))).))))))...((.(.((((((------...))))--))).))......... ( -26.30) >DroYak_CAF1 1048 87 + 1 GCUUGUGCACGAGGAUCUCUGCAAGAAGUGCAAUUCUCUUCUCGAGCACCAUAACGAGUAAGUUU------UACGAGC--CACGUGACCUGAAAU ((((((((.((((((....((((.....))))......)))))).))))........(((.....------)))))))--............... ( -18.80) >consensus GCUGGUGCACGAGGAUCUCUGCAAGAAGUGCAAUUCACUUCUCGAGCACCAUAACGAGUAAGUUG______CACAAUC__CACGCAACCUGAAAU ..((((((.((((((....((((.....))))......)))))).))))))............................................ (-16.29 = -16.82 + 0.53)


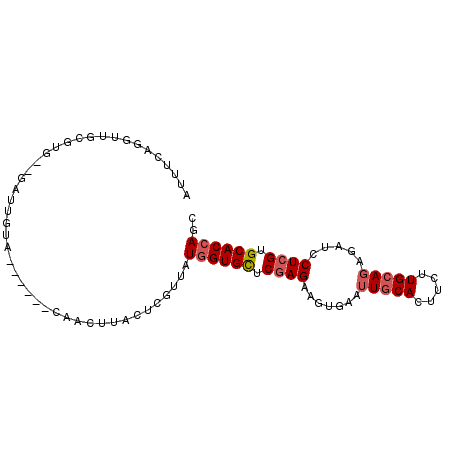
| Location | 8,826,638 – 8,826,733 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -15.79 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8826638 95 - 27905053 AUUUCAGAUUGGGUGCACAAUCUACCCAAUCUACUUACUCGUUAUGGUGCUCGAGAAGUGAAUUGCACUUCUUGCAGAGAUCCUCGUGCACCAGC .....((((((((((.......))))))))))............((((((.(((((((((.....)))))))))..(((...)))..)))))).. ( -34.90) >DroVir_CAF1 1531 85 - 1 GCUGCA-GUUAUAUC--GA-UGAA------UCACUCACUCAUUAUGGUGUUCCAGCAUUGCAUUACACUUCUUGCAUAGGUCCUCGUGCACCAGC ...((.-........--((-(((.------........))))).((((((....((..((((..........))))...))......)))))))) ( -14.30) >DroSec_CAF1 1105 87 - 1 AUUUCAGGUUGCGUA--GAUUGUG------CAACUUACUCGUUAUGGUGCUCGAGAAGUGAAUUGCACUUCUUGCAGAGAUCCUCGUGCACCAGC .....((((((((..--.....))------))))))........((((((.(((((((((.....)))))))))..(((...)))..)))))).. ( -29.40) >DroSim_CAF1 1062 87 - 1 AUUUCAGGUUGCGCA--GAUUGUG------CAACUUACUCGUUAUGGUGCUCGAGAAGUGAAUUGCACUUCUUGCAGAGAUCCUCGUGCACCAGC .....(((((((((.--....)))------))))))........((((((.(((((((((.....)))))))))..(((...)))..)))))).. ( -33.60) >DroEre_CAF1 1130 87 - 1 AUUUCAGGUCGCGAG--GCUCGUG------AAACCUACUCGUUAUGGUGCUCGAGAAGAGAAUUGCAUUUCUUGCAGAGAUCCUCGUGCACCAGC .(((((((((....)--)))..))------)))...........((((((.((((..((...(((((.....)))))...)))))).)))))).. ( -29.90) >DroYak_CAF1 1048 87 - 1 AUUUCAGGUCACGUG--GCUCGUA------AAACUUACUCGUUAUGGUGCUCGAGAAGAGAAUUGCACUUCUUGCAGAGAUCCUCGUGCACAAGC ...........((((--((..(((------.....)))..))))))((((.((((..((...(((((.....)))))...)))))).)))).... ( -22.10) >consensus AUUUCAGGUUGCGUG__GAUUGUA______CAACUUACUCGUUAUGGUGCUCGAGAAGUGAAUUGCACUUCUUGCAGAGAUCCUCGUGCACCAGC ............................................((((((.((((.......(((((.....))))).....)))).)))))).. (-15.79 = -16.32 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:50 2006