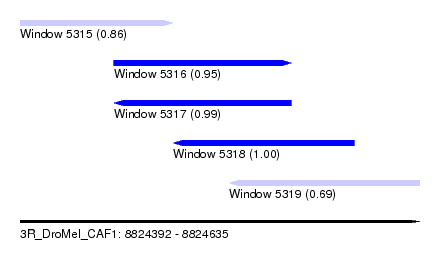
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,824,392 – 8,824,635 |
| Length | 243 |
| Max. P | 0.999427 |

| Location | 8,824,392 – 8,824,485 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8824392 93 + 27905053 UUCCUGCUGCCAUGUGAUGUCUCUGUCCUCCAUGGACGUAGAUGUAGAU------GCGGAUGC------GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUG ......((((.((.(.((((((.(((((.....))))).))))))).))------))))....------((((((((.(((((...))))).))))))))..... ( -38.40) >DroSec_CAF1 3000 87 + 1 UUCCUGCUGCCAUGUGAUGUCUCUGUCCUCCAUGGAUG------UAGAU------GUGGAUGC------GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUG .....((..((((.....((((..((((.....)))).------.))))------))))..))------((((((((.(((((...))))).))))))))..... ( -34.90) >DroSim_CAF1 2999 87 + 1 UUCCUGCUGCCAUGUGAUGUCUCUGUCCUCCAUGGAUG------UAGAU------GUGGAUGC------GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUG .....((..((((.....((((..((((.....)))).------.))))------))))..))------((((((((.(((((...))))).))))))))..... ( -34.90) >DroYak_CAF1 2972 99 + 1 UUCCUGCUGCCAUGUGAUGUCCCUGUCCUCCAUGGAUG------UAGAUGCUGAUGUGGAUGCGGAUGCGGAGGGCGAUGUGGAUGCCGCAACGUCCUCCUCCUG ...((((..((((((.(((((..(((((.....)))))------..)))).).))))))..))))....((((((((.(((((...))))).))))))))..... ( -39.40) >consensus UUCCUGCUGCCAUGUGAUGUCUCUGUCCUCCAUGGAUG______UAGAU______GUGGAUGC______GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUG .....((..((((.....((((.(((((.....))))).......))))......))))..))......((((((((.(((((...))))).))))))))..... (-29.33 = -29.45 + 0.12)


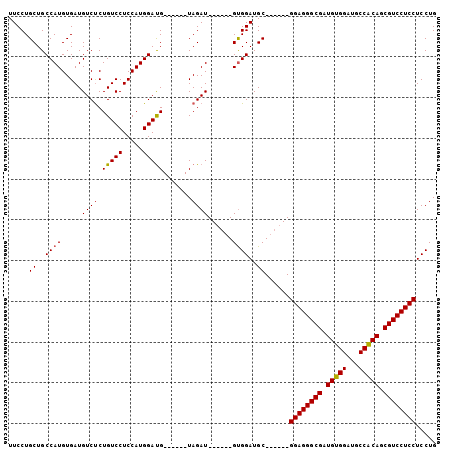
| Location | 8,824,449 – 8,824,557 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -50.86 |
| Consensus MFE | -47.34 |
| Energy contribution | -48.02 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8824449 108 + 27905053 ----GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUGCGUCGCCACCGCUUCCCAGCUGAAUCCGUCGCCACCGCCUGCUGGUGUCCAACAGGCGAAGCCGGCGCGGAG ----((((((((.(((((...))))).))))))))..((((((((.....(((((...((.((.....))))....(((((.(((...))).)))))))))))))))))).. ( -51.60) >DroSec_CAF1 3051 108 + 1 ----GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUGCGUCGCCACCGCCACCCAGCUGAAUUCGUCGCCACCGCCUGCUGGUGUCCAACAGGCGAAGCCGGCGCGGAG ----((((((((.(((((...))))).))))))))..((((((((.(..((((..(((((.((.....))((....))..)))))((.....))))))..).)))))))).. ( -49.50) >DroSim_CAF1 3050 108 + 1 ----GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUGCGUCGCCACCGCUUCCCAGCUGAAUUCGUCGCCACCGCCUGCUGGUGUCCAACAGGCGAAGCCGGCGCGGAG ----((((((((.(((((...))))).))))))))..((((((((.....(((((...((.((.....))))....(((((.(((...))).)))))))))))))))))).. ( -51.60) >DroEre_CAF1 3165 109 + 1 AUG---AGGGCGAUGUGGAUGCCACAACGUCCUCCUCCUGCGGUGCCACCGCUUCCCAGCUGAAUCCGUCGCCACCGCCUGCUGGUGUCCAACAGGCGAAGCCGGCGCGGAG ..(---((((((.(((((...))))).))))))).....((((.....))))............((((.((((..((((((.(((...))).)))))).....)))))))). ( -50.10) >DroYak_CAF1 3031 112 + 1 AUGCGGAGGGCGAUGUGGAUGCCGCAACGUCCUCCUCCUGCGUCGCCACCGCUUCCCAGCUGAAUCCGUCGCCACCGCCUGCUGGUGUCCAACAGGCGAAGCCGGCGCGGAG ....((((((((.(((((...))))).))))))))..((((((((.....(((((...((.((.....))))....(((((.(((...))).)))))))))))))))))).. ( -51.50) >consensus ____GGAGGGCGAUGUGGAUGCCACAGCGUCCUCCUCCUGCGUCGCCACCGCUUCCCAGCUGAAUCCGUCGCCACCGCCUGCUGGUGUCCAACAGGCGAAGCCGGCGCGGAG ....((((((((.(((((...))))).))))))))..((((((((.....(((((...((.((.....))))....(((((.(((...))).)))))))))))))))))).. (-47.34 = -48.02 + 0.68)


| Location | 8,824,449 – 8,824,557 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -54.48 |
| Consensus MFE | -50.92 |
| Energy contribution | -51.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8824449 108 - 27905053 CUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCGACGCAGGAGGAGGACGCUGUGGCAUCCACAUCGCCCUCC---- .(((((((((...(((((((((((...)))))))))))))))).))))(((.(((....))).(((....))).)))(((((.((.(((((...))))).)).)))))---- ( -55.80) >DroSec_CAF1 3051 108 - 1 CUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGAAUUCAGCUGGGUGGCGGUGGCGACGCAGGAGGAGGACGCUGUGGCAUCCACAUCGCCCUCC---- (.((((.(((((((((((((((((...)))))))))))((....)).....)))))))))).)(((....)))....(((((.((.(((((...))))).)).)))))---- ( -53.80) >DroSim_CAF1 3050 108 - 1 CUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGAAUUCAGCUGGGAAGCGGUGGCGACGCAGGAGGAGGACGCUGUGGCAUCCACAUCGCCCUCC---- ((((((.(((((((((((((((((...)))))))))))((....)).....))))))...)).(((....))).))))((((.((.(((((...))))).)).)))).---- ( -53.30) >DroEre_CAF1 3165 109 - 1 CUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCACCGCAGGAGGAGGACGUUGUGGCAUCCACAUCGCCCU---CAU ((((...(((((((((((((((((...)))))))))))((....)).....))))))...((((.....)))).))))((((.((.(((((...))))).)).)))---).. ( -55.40) >DroYak_CAF1 3031 112 - 1 CUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCGACGCAGGAGGAGGACGUUGCGGCAUCCACAUCGCCCUCCGCAU .(((((((((...(((((((((((...)))))))))))))))).))))....((.(((..(((((((((((((........).))))))((...)).))))))..))))).. ( -54.10) >consensus CUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCGACGCAGGAGGAGGACGCUGUGGCAUCCACAUCGCCCUCC____ ((((((.(((((((((((((((((...)))))))))))((....)).....))))))...)).(((....))).))))((((.((.(((((...))))).)).))))..... (-50.92 = -51.32 + 0.40)


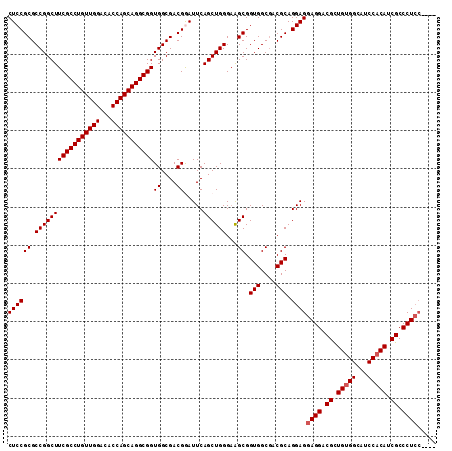
| Location | 8,824,485 – 8,824,595 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -58.62 |
| Consensus MFE | -53.34 |
| Energy contribution | -54.42 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8824485 110 - 27905053 AGCUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCGACG .(((..((((((((...((((((....((((....--))))(((((((((...(((((((((((...)))))))))))))))).)))).)))))).)).))))))))).... ( -60.20) >DroSec_CAF1 3087 110 - 1 AGUUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGAAUUCAGCUGGGUGGCGGUGGCGACG .((((.((((((((...((((((...(((((....--))))).(((((((...(((((((((((...)))))))))))))))).))...)))))).)).))))))..)))). ( -57.50) >DroSim_CAF1 3086 112 - 1 AGUUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGCCGUGUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGAAUUCAGCUGGGAAGCGGUGGCGACG .((((.((((((((.(((((..(((((((...))))))))))))((((((...(((((((((((...)))))))))))))))).)...........)).))))))..)))). ( -62.00) >DroEre_CAF1 3202 110 - 1 AGUUGAGCCGCUCCAGGAGAUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCACCG ......((((..(((.....)))..))))((((.(--(((((((((((((...(((((((((((...)))))))))))))))).))))(((......))))))))..)))). ( -55.50) >DroYak_CAF1 3071 110 - 1 AGUUGAGCCGCUCCAGGAGAUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCGACG .((((.((((((((.....((.((...)).)).((--(((.(((((((((...(((((((((((...)))))))))))))))).))))..))))).)).))))))..)))). ( -57.90) >consensus AGUUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGAG__CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGGUGGCGACGGAUUCAGCUGGGAAGCGGUGGCGACG .((((.((((((((...((((((....((((......))))(((((((((...(((((((((((...)))))))))))))))).)))).)))))).)).))))))..)))). (-53.34 = -54.42 + 1.08)



| Location | 8,824,519 – 8,824,635 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -52.10 |
| Consensus MFE | -49.60 |
| Energy contribution | -50.64 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8824519 116 - 27905053 AGGAUCGUGUGCAGCGUGUUGAUGAUGUUGCCGUCGGGAAAGCUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGG .(((.((.((.((((...((..(((((....)))))..)).)))).)))).))).(((((((((..(((((....--)))))....)))))))))(((((((((...))))))))).. ( -56.20) >DroSec_CAF1 3121 116 - 1 AGGAUCGUGUGCAGCGUGUUGAUGAUGUUGCCGUCGGGAAAGUUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGG .(((.((.((.((((...((..(((((....)))))..)).)))).)))).))).(((((((((..(((((....--)))))....)))))))))(((((((((...))))))))).. ( -53.80) >DroSim_CAF1 3120 118 - 1 AGGAUCGUGUGCAGCGUGUUGAUGAUGUUGCCGUCGGGAAAGUUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGCCGUGUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGG .(((.((.((.((((...((..(((((....)))))..)).)))).)))).))).(((((((((...((((.((.....)).)))))))))))))(((((((((...))))))))).. ( -56.70) >DroEre_CAF1 3236 116 - 1 AGGAUCGUGUGCAGCGUGUUAAUGAUAUUGCCCUCGGGAAAGUUGAGCCGCUCCAGGAGAUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGG .((..((((.(((((...(((((.......((....))...)))))(((((((((.....)))...))))))..)--))))..)))))).....((((((((((...)))))))))). ( -46.90) >DroYak_CAF1 3105 116 - 1 AGGAUCGUGUGCAGCGUGUUAAUGAUAUUGCCCUCGGGAAAGUUGAGCCGCUCCAGGAGAUGGCAUGGCGGUGAG--CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGG .((..((((.(((((...(((((.......((....))...)))))(((((((((.....)))...))))))..)--))))..)))))).....((((((((((...)))))))))). ( -46.90) >consensus AGGAUCGUGUGCAGCGUGUUGAUGAUGUUGCCGUCGGGAAAGUUGAGCCGCUCCAGGAGCUGGCAUGGCGGUGAG__CUGCUCCGCGCCGGCUUCGCCUGUUGGACACCAGCAGGCGG .(((.((.((.((((...((..(((((....)))))..)).)))).)))).))).(((((((((...((((.(.......).)))))))))))))(((((((((...))))))))).. (-49.60 = -50.64 + 1.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:48 2006