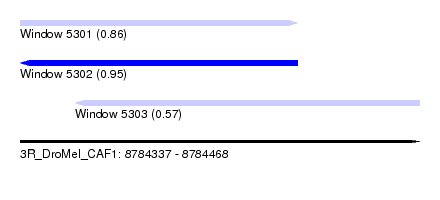
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,784,337 – 8,784,468 |
| Length | 131 |
| Max. P | 0.946268 |

| Location | 8,784,337 – 8,784,428 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8784337 91 + 27905053 CCGGCAACAUCAUUUGCUGCCUGCAUUAGUGGCAAAAAUGUGAAUUCAAUCCCAGUGAAUUGUUGCCUUUAAUGCGAUUAAAAGUGACAGA ..((((.((.....)).))))((((((((.(((((.......((((((.......)))))).))))).))))))))............... ( -22.80) >DroSim_CAF1 222790 91 + 1 CCGGCAACAUCAUUUGCUGCCUGCAUUACUGGCCUAAAUGUGAAUUCCAUCCCGGGGAAUUGUUGCCUUUAAUGCAAUAAAAAUUGGCAAA ((((((.((.....)).))))(((((((..(((...(((...((((((.......))))))))))))..))))))).........)).... ( -23.20) >DroEre_CAF1 222873 90 + 1 CCGGCAACAUCAUUUGCUGGCUGCAUUAGUGGCAGGAAUGUGAAUUCAAUCC-GGUGAAUUGUUGCCUUUAAUGCGAUUAAGGGGGGCAGA (((((((......)))))))((((.(((((.((((((.((......)).)))-((..(....)..)).....))).))))).....)))). ( -29.00) >DroYak_CAF1 216194 90 + 1 CCGGCAACAUAAUUUGCUGGCUGCAUUAGUGGCAGAAAUGUGAAUUCAAUCC-UGUGAAUUGCUGCCUUUAAUGCGAUUAAGAGGGGCAGA (((((((......)))))))((((.....(.(((....))).)......(((-(.(.((((((..........)))))).).)))))))). ( -27.20) >consensus CCGGCAACAUCAUUUGCUGCCUGCAUUAGUGGCAGAAAUGUGAAUUCAAUCC_GGUGAAUUGUUGCCUUUAAUGCGAUUAAAAGGGGCAGA .((((((......))))))..((((((((.(((((.......((((((.......)))))).))))).))))))))............... (-21.56 = -21.50 + -0.06)



| Location | 8,784,337 – 8,784,428 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.95 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8784337 91 - 27905053 UCUGUCACUUUUAAUCGCAUUAAAGGCAACAAUUCACUGGGAUUGAAUUCACAUUUUUGCCACUAAUGCAGGCAGCAAAUGAUGUUGCCGG ................((((((..(((((((((((....))))))...........)))))..)))))).(((((((.....))))))).. ( -26.40) >DroSim_CAF1 222790 91 - 1 UUUGCCAAUUUUUAUUGCAUUAAAGGCAACAAUUCCCCGGGAUGGAAUUCACAUUUAGGCCAGUAAUGCAGGCAGCAAAUGAUGUUGCCGG ....((.........(((((((..(((...((((((.......)))))).........)))..)))))))(((((((.....))))))))) ( -27.00) >DroEre_CAF1 222873 90 - 1 UCUGCCCCCCUUAAUCGCAUUAAAGGCAACAAUUCACC-GGAUUGAAUUCACAUUCCUGCCACUAAUGCAGCCAGCAAAUGAUGUUGCCGG ................((((((..((((..........-(((.((......)).)))))))..))))))..((.((((......)))).)) ( -19.00) >DroYak_CAF1 216194 90 - 1 UCUGCCCCUCUUAAUCGCAUUAAAGGCAGCAAUUCACA-GGAUUGAAUUCACAUUUCUGCCACUAAUGCAGCCAGCAAAUUAUGUUGCCGG ................((((((..((((((((((....-.)))))...........)))))..))))))..((.((((......)))).)) ( -20.70) >consensus UCUGCCACUCUUAAUCGCAUUAAAGGCAACAAUUCACC_GGAUUGAAUUCACAUUUCUGCCACUAAUGCAGCCAGCAAAUGAUGUUGCCGG ....((..........((((((..(((((((((((....))))))...........)))))..)))))).(((((((.....))))))))) (-20.76 = -21.95 + 1.19)


| Location | 8,784,355 – 8,784,468 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.04 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -13.95 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8784355 113 - 27905053 GCUAGGCUACAAUUAUUUGCCAAGUUGGCAAGUUGGCCUUUCUGUCACUUUUAAUCGCAUUAAAGGCAACAAUUCACUGGGAUUGAAUUCACAUUUUUGCCACUAAUGCAGGC (((((((((.....(((((((.....))))))))))))).................((((((..(((((((((((....))))))...........)))))..)))))).))) ( -31.80) >DroSim_CAF1 222808 103 - 1 UCGACGAUCCAAUUAGUUGCCAAGU----------ACCUUUUUGCCAAUUUUUAUUGCAUUAAAGGCAACAAUUCCCCGGGAUGGAAUUCACAUUUAGGCCAGUAAUGCAGGC ........((....(((((.((((.----------.....)))).))))).....(((((((..(((...((((((.......)))))).........)))..))))))))). ( -20.30) >DroEre_CAF1 222891 102 - 1 GCGAGGCUGCAAUUAGUUGCCAACU----------UUCUUUCUGCCCCCCUUAAUCGCAUUAAAGGCAACAAUUCACC-GGAUUGAAUUCACAUUCCUGCCACUAAUGCAGCC ....(((((((.(((((.((.....----------.......((((....(((((...))))).))))..........-(((.((......)).))).)).)))))))))))) ( -25.90) >DroYak_CAF1 216212 92 - 1 ----------AAUUAGUUGUCAACA----------UUCUUUCUGCCCCUCUUAAUCGCAUUAAAGGCAGCAAUUCACA-GGAUUGAAUUCACAUUUCUGCCACUAAUGCAGCC ----------...............----------.....................((((((..((((((((((....-.)))))...........)))))..)))))).... ( -16.30) >consensus GCGAGGCU_CAAUUAGUUGCCAACU__________UCCUUUCUGCCACUCUUAAUCGCAUUAAAGGCAACAAUUCACC_GGAUUGAAUUCACAUUUCUGCCACUAAUGCAGCC ........................................................((((((..(((((((((((....))))))...........)))))..)))))).... (-13.95 = -14.83 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:31 2006