| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,769,080 – 8,769,185 |
| Length | 105 |
| Max. P | 0.708357 |

| Location | 8,769,080 – 8,769,185 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
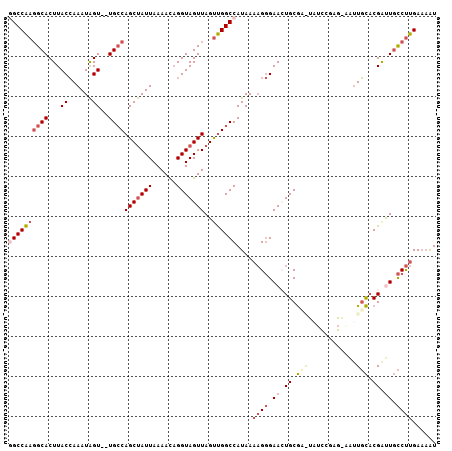
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -29.68 |
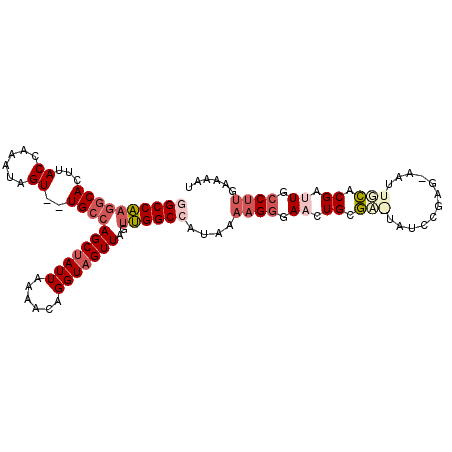
| Consensus MFE | -17.35 |
| Energy contribution | -19.02 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8769080 105 + 27905053 GGCCAAGGCACUUACCAAAUAGU--UGCCAGCUAUUAAAACAGGUAGUUAGUUGGCCAUAAAAGGGAACUGCGCGUAUCCGAGAAAUUGCACGAUUGCCUUGAAAAU ((((((((((((........)).--))))(((((((......)))))))..))))))....((((.((.((.(((............))).)).)).))))...... ( -29.00) >DroPse_CAF1 209777 103 + 1 UGCCG-CGCACUUACAAAAUGGUGGUGGCAGCCAUUAAAAUUGGUAGUUAGCCGGCCAUAAA-GCGAACUGCAAAUGGUUUAAGCGCUUUACGACUCUCCCGA--GU (((((-(.((((........))))))))))((((.......)))).((.((((.(((((...-((.....))..)))))....).)))..)).((((....))--)) ( -28.50) >DroSec_CAF1 189848 95 + 1 GGCCAAGGCACUUACCAAAUAGU--UGCCAGCUAUUAAAACAGGUAGUUAGUUGGCCAUAAAAGGGAACUGCGC----------AAUUGCACGAUUGCCUUGAAAAU ((((((((((((........)).--))))(((((((......)))))))..))))))....((((.((.((.((----------....)).)).)).))))...... ( -29.40) >DroSim_CAF1 207237 95 + 1 GGCCAAGGCACUUACCAAAUAGU--UGCCAGCUAUUAAAACAGGUAGUUAGUUGGCCAUAAAAGGGAACUGCGC----------AAUUGCACGAUUGCCUUGAAAAU ((((((((((((........)).--))))(((((((......)))))))..))))))....((((.((.((.((----------....)).)).)).))))...... ( -29.40) >DroEre_CAF1 206716 105 + 1 AGCCAAGGCACUUACCAAAUAGU--UGCCAGCUAUUAAAACAGGUAGUUAGUUGGCCAUAAAAGGGAAGUGCGAGUAUCCGAGGAAUUGCCCGAUUGCCUUGAAAAU ...((((((((((.((.....((--.((((((((..............)))))))).)).....))))))))..(((..((.((.....))))..)))))))..... ( -30.24) >DroYak_CAF1 199579 105 + 1 GGCCAAGGCACUUACCAAAUAGU--UGCCAGCUAUUAAAACAGGUAGUUAGUUGGCCAUAAAAGGGAAGUGCGAAUAUCCGAGCAAUUGCACGGCUGCCUUGAAAAU (((....((((((.((.....((--.((((((((..............)))))))).)).....))))))))......(((.((....)).)))..)))........ ( -31.54) >consensus GGCCAAGGCACUUACCAAAUAGU__UGCCAGCUAUUAAAACAGGUAGUUAGUUGGCCAUAAAAGGGAACUGCGA_UAUCCGAG_AAUUGCACGAUUGCCUUGAAAAU ((((((((((...((......))..))))(((((((......)))))))..))))))....((((.((.((.(((............))).)).)).))))...... (-17.35 = -19.02 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:23 2006