
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,756,521 – 8,756,628 |
| Length | 107 |
| Max. P | 0.860183 |

| Location | 8,756,521 – 8,756,628 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8756521 107 - 27905053 AUUGUGGAUUGAUGCUCAUUAAUCCCACAAAUGCCU---CAA--AACAUGUAAAAAGCGGAAUCCACAACAAAAAGUGUUCAAGUGGAAAACUUGUCCAGCGCACACACACA----- .(((((((((..((((..(((.........(((...---...--..))).)))..)))).)))))))))......((((((((((.....)))))...))))).........----- ( -22.10) >DroVir_CAF1 220108 107 - 1 AUUGUAGAUUGUUGCUCAUUAAUCCCUCGAAUACAUAUUGAAUCUAUCUGUAGAA----AAAUCUACAACAA-AAGUGUUCAAGUGGAAAACUUGUUCAGCGCUGAACAAC--A--- ......................(((((.((((((.....((.....))((((((.----...))))))....-..)))))).)).)))....(((((((....))))))).--.--- ( -21.80) >DroGri_CAF1 180696 96 - 1 AUUGUAGAUUGUUGCUCAUUAAUCCCUCAAA-----------UCGAUGUGUAUAA----AAAUCUACAACAA-AAGUGUUCAAGUGGAAAACUUGUUCAGCACUGAACAAC--A--- .(((((((((..(((.(((..((.......)-----------)..))).)))...----.)))))))))...-.(((((((((((.....)))))...)))))).......--.--- ( -23.00) >DroSim_CAF1 194874 112 - 1 AUUGUGGAUUGAUGCUCAUUAAUCCCACAAAUGCCU---CAA--AACAUGUAAAAAGCGGAAUCCACAACAAAAAGUGUUCAAGUGGAAAACUUGUCCAGCGCACACAAACACAGAC .(((((((((..((((..(((.........(((...---...--..))).)))..)))).)))))))))......((((((((((.....)))))...))))).............. ( -22.10) >DroWil_CAF1 220458 104 - 1 GUGCCGAAUUGAUGCUCAUUAAUCCCUCAAAUACUU---GAA--AACAUGUAAAAAAAGAAAUCGACUACUA-GAGUGUUCGAGUGGA-AACUUGUCCAU---ACACACACACA--- (((........(((..((....((((((.(((((((---...--............................-))))))).))).)))-....))..)))---.......))).--- ( -15.64) >DroAna_CAF1 167204 104 - 1 AUUGUGGAUUGAUGCUCAUUAAUCCCACAAAUGCCU---CGA--AACAUGUAAAAAGCGGAAUCCACAACAAAAAGUGUUCGAGUGGAAAACUUGUCCAACACACACAC-------- .(((((((((..((((..(((.........(((...---...--..))).)))..)))).)))))))))......((((((((((.....)))))...)))))......-------- ( -22.20) >consensus AUUGUGGAUUGAUGCUCAUUAAUCCCACAAAUGCCU___CAA__AACAUGUAAAAAGCGAAAUCCACAACAA_AAGUGUUCAAGUGGAAAACUUGUCCAGCGCACACAAAC__A___ .(((((((((..................................................)))))))))......((((((((((.....)))))...))))).............. (-11.80 = -12.03 + 0.23)
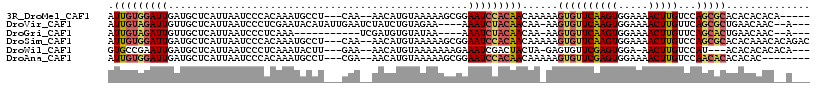

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:18 2006