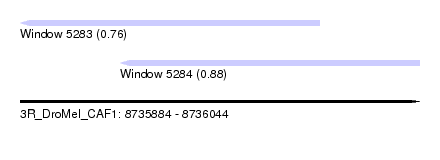
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,735,884 – 8,736,044 |
| Length | 160 |
| Max. P | 0.878937 |

| Location | 8,735,884 – 8,736,004 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -25.69 |
| Energy contribution | -24.78 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8735884 120 - 27905053 GAGAAGCAGCUGGAACCAAAGGAAAUGGUUCUAUUGGAGCCCUUUAAGGCCGGUUACGCCUUUGUUGUGGGCGUGUACAUUCGUAAGGAUGUUUCCUUGUAGUUGUCAUUAUACAUGUUU .....(((((((((((((.......)))))))...(((((((...(((((.(....))))))......))))....((((((....)))))).)))....)))))).............. ( -36.50) >DroSec_CAF1 162484 120 - 1 GAGAAGCAGCUGGAACCAAAAGAAAUGGUUCUAUUGGAGCCCUUUAAGGCCGGUUACGCCUUUGUUGUGGGCGUGUACACACGUAAGGAUGUUUCCUUGUAGAUAUAAUUAUACAUGUUU ......(((.((((((((.......)))))))))))..((((...(((((.(....))))))......))))(((....))).((((((....))))))..................... ( -32.10) >DroSim_CAF1 179661 120 - 1 GAGAAGCAGCUGGAACCAAAAGAAAUGGUUCUAUUGGAGCCCUUUAAGGCCGGUUACGCCUUUGUUGUGGGCGUGUACACACGUAAGGAUGUUACCUUGUAGAUAUAAUUAUACACGUUU ......(((.((((((((.......)))))))))))..((((...(((((.(....))))))......))))(((((((..(....)..))((((...)))).........))))).... ( -30.30) >DroEre_CAF1 178759 117 - 1 GAGCAGCAGUUGGAGCCAAUCGAAAUGGUUCUAUUGGAGCCCUUUAAGCCCGGCUACGCCCUUGUUGUGGGCGUGUACACACGUAAGGAUGCUGCCUUGUAGUUAUC---UUAUAUAUAU ..(((((((((((.((......(((.(((((.....))))).)))..)))))))(((((((.......))))(((....))))))....))))))............---.......... ( -35.90) >DroYak_CAF1 171121 120 - 1 GAGCAGCAGUUGGAACCAAUCGAAAUGGUUCUAUUGGAGCCCUUUAAGCCCGGCUACGCCCUUGUUGUGGGCGUGUAUACACGUAAGGAUGCUGCUUUGUAGUUGUUAUGGUAUAUGUUU (((((((...((((((((.......)))))))).(((.((.......))))))))((((((.......))))))((((((.(((((.(((((......))).)).))))))))))))))) ( -38.00) >DroPer_CAF1 173764 104 - 1 AAAUACCAUCUUGAGCCUCUCCAGCUGGUCCAACUGGAUCCCUAUGUUCGUGGUAAUGCUCUGGUAGUGGGCGUUUACACUCGAUUGC---CAGUGUUAUAAUGAUC------------- ...((((((....(((.......)))(((((....))))).........))))))..((.(((((((((((.(....).))).)))))---))).))..........------------- ( -27.20) >consensus GAGAAGCAGCUGGAACCAAACGAAAUGGUUCUAUUGGAGCCCUUUAAGCCCGGUUACGCCCUUGUUGUGGGCGUGUACACACGUAAGGAUGCUGCCUUGUAGUUAUCAUUAUACAUGUUU ..((((((..((((((((.......)))))))).......((((..........(((((((.......))))))).........)))).))))))......................... (-25.69 = -24.78 + -0.91)



| Location | 8,735,924 – 8,736,044 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -30.73 |
| Energy contribution | -30.23 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8735924 120 - 27905053 UAAUAAGGAUUCCUUUGCCGCCGAGAGGAGACUUUUAAAGGAGAAGCAGCUGGAACCAAAGGAAAUGGUUCUAUUGGAGCCCUUUAAGGCCGGUUACGCCUUUGUUGUGGGCGUGUACAU ....((((...)))).(((((((((((....)))))(((((.(...(((.((((((((.......)))))))))))...))))))..)).))))(((((((.......)))))))..... ( -38.70) >DroPse_CAF1 178865 120 - 1 CACGAAGAUGGCCCUCGAAGCCGAGAUGGAGCUGCUCAAGAAAUACCAUCUUGAGCCUCUCCAGCUGGUCCAACUGGAUCCCUAUGUUCGUGGCAAUGCUCUGGUAGUGGGCGUUUACAC ((((((.((((..((((....)))).(((((..((((((((.......))))))))..)))))...(((((....))))).)))).))))))..(((((((.......)))))))..... ( -47.20) >DroSec_CAF1 162524 120 - 1 UAAUAAGGAUUCCUUUGCCGCCGAGAGGAGACUCUUAAAGGAGAAGCAGCUGGAACCAAAAGAAAUGGUUCUAUUGGAGCCCUUUAAGGCCGGUUACGCCUUUGUUGUGGGCGUGUACAC ....((((...)))).(((((((((((....)))))(((((.(...(((.((((((((.......)))))))))))...))))))..)).))))(((((((.......)))))))..... ( -41.30) >DroSim_CAF1 179701 120 - 1 UAAUAAGGAUUCCUUUGCCGCCGAGAGGAGACUCUUAAAGGAGAAGCAGCUGGAACCAAAAGAAAUGGUUCUAUUGGAGCCCUUUAAGGCCGGUUACGCCUUUGUUGUGGGCGUGUACAC ....((((...)))).(((((((((((....)))))(((((.(...(((.((((((((.......)))))))))))...))))))..)).))))(((((((.......)))))))..... ( -41.30) >DroEre_CAF1 178796 120 - 1 UAAUAAGGAUACCUUUGCCGCCGAGAGGAAACUGUUGAAGGAGCAGCAGUUGGAGCCAAUCGAAAUGGUUCUAUUGGAGCCCUUUAAGCCCGGCUACGCCCUUGUUGUGGGCGUGUACAC ...(((((....)))))..((((..((....)).(((((((.((..((((.(((((((.......)))))))))))..)))))))))...))))(((((((.......)))))))..... ( -43.20) >DroYak_CAF1 171161 120 - 1 CAAUAAGGAUACCUUUGCCGCCGAGAGGAAACUUUUGAAAGAGCAGCAGUUGGAACCAAUCGAAAUGGUUCUAUUGGAGCCCUUUAAGCCCGGCUACGCCCUUGUUGUGGGCGUGUAUAC ........((((....((((.((((((....))))))((((.((..((((.(((((((.......)))))))))))..)).)))).....)))).((((((.......)))))))))).. ( -42.10) >consensus UAAUAAGGAUUCCUUUGCCGCCGAGAGGAAACUCUUAAAGGAGAAGCAGCUGGAACCAAACGAAAUGGUUCUAUUGGAGCCCUUUAAGCCCGGCUACGCCCUUGUUGUGGGCGUGUACAC ...................((((..((....))((((((((.(...(((.((((((((.......)))))))))))...)))))))))..))))(((((((.......)))))))..... (-30.73 = -30.23 + -0.49)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:12 2006