
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,691,801 – 8,691,920 |
| Length | 119 |
| Max. P | 0.994358 |

| Location | 8,691,801 – 8,691,920 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.15 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
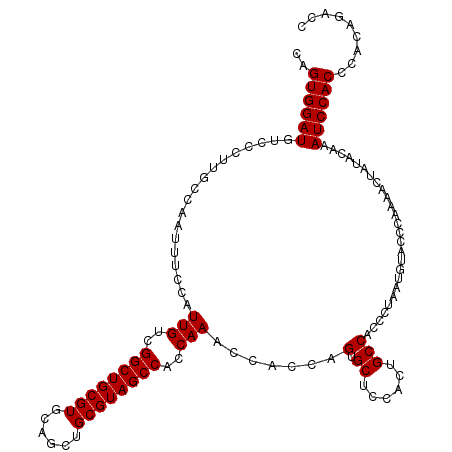
>3R_DroMel_CAF1 8691801 119 + 27905053 CAGUGGAUGUCCCUUGCCAAUUUCCAUUGUCGGCUGCGUGCAGCUGCGUAGCCACCAAGCCACCAGUGCUCCACUGCCACCCUAAUGUACCCAAAACUGCACAAAUCCACCCACAGACC ..((((((......(((.........(((..((((((((......))))))))..))).....(((((...)))))......................)))...))))))......... ( -28.70) >DroSec_CAF1 118328 119 + 1 CAGUGGAUGUCCCUUGCCAAUUUCCAUUGUCGGCUGCGUGCAGCUGCGUAGCCACCAAACCACCAGCGCUCCACUGCCACCCUAAUGUACCCAAAACUAUACAAAUCCACCCACAGACC ((((((((((.....(.((((....)))).)((((((((......))))))))............))).)))))))........................................... ( -25.80) >DroSim_CAF1 134906 119 + 1 CAGUGGAUGUCCCUUGCCAAUUUCCAUUGUCGGCUGCGUGCAGCUGCGUAGCCACCAAACCACCAGCGCUCCACUGCCACCCUAAUGUACCCAAAACUAUACAAAUCCACCCACAGACC ((((((((((.....(.((((....)))).)((((((((......))))))))............))).)))))))........................................... ( -25.80) >DroEre_CAF1 132231 108 + 1 CAGUGGAUGUCCCUUGCCAAUUUCUAUUGUCGGCUGCGUGCAGCUGCGUAGCCACCAAACCACCAGUGCUCCA--GCCACCCUAAUGUACCCAAAACUUUCCAAAUCCAC--------- ..((((((.......(.((((....)))).)(((((.(.(((.(((.((.(........).)))))))).)))--)))..........................))))))--------- ( -24.70) >DroYak_CAF1 124716 113 + 1 CAGUGGAUGUCCCUUGCCAAUUUCUAUUGUCGGCUGCGUGCAGCUGCGUAGCCACCAAACCACCAGUGCUCCA--GCCACCCUAAUAUACCCAAAACUAUCCAAAUCCACCCACA---- ..((((((.......(.((((....)))).)(((((.(.(((.(((.((.(........).)))))))).)))--)))..........................)))))).....---- ( -25.50) >consensus CAGUGGAUGUCCCUUGCCAAUUUCCAUUGUCGGCUGCGUGCAGCUGCGUAGCCACCAAACCACCAGUGCUCCACUGCCACCCUAAUGUACCCAAAACUAUACAAAUCCACCCACAGACC ..((((((..................(((..((((((((......))))))))..))).......(.((......)))..........................))))))......... (-24.40 = -24.40 + 0.00)

| Location | 8,691,801 – 8,691,920 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.15 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -34.26 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8691801 119 - 27905053 GGUCUGUGGGUGGAUUUGUGCAGUUUUGGGUACAUUAGGGUGGCAGUGGAGCACUGGUGGCUUGGUGGCUACGCAGCUGCACGCAGCCGACAAUGGAAAUUGGCAAGGGACAUCCACUG .....(((((((..(((((((((((.(((((.(((((((.((.(((((...))))).)).)))))))))).)).))))))))...(((((.........))))).)))..))))))).. ( -45.50) >DroSec_CAF1 118328 119 - 1 GGUCUGUGGGUGGAUUUGUAUAGUUUUGGGUACAUUAGGGUGGCAGUGGAGCGCUGGUGGUUUGGUGGCUACGCAGCUGCACGCAGCCGACAAUGGAAAUUGGCAAGGGACAUCCACUG .....(((((((..(((((.....................((((.(((..((((((((((((....)))))).)))).)).))).)))).((((....)))))))))...))))))).. ( -41.50) >DroSim_CAF1 134906 119 - 1 GGUCUGUGGGUGGAUUUGUAUAGUUUUGGGUACAUUAGGGUGGCAGUGGAGCGCUGGUGGUUUGGUGGCUACGCAGCUGCACGCAGCCGACAAUGGAAAUUGGCAAGGGACAUCCACUG .....(((((((..(((((.....................((((.(((..((((((((((((....)))))).)))).)).))).)))).((((....)))))))))...))))))).. ( -41.50) >DroEre_CAF1 132231 108 - 1 ---------GUGGAUUUGGAAAGUUUUGGGUACAUUAGGGUGGC--UGGAGCACUGGUGGUUUGGUGGCUACGCAGCUGCACGCAGCCGACAAUAGAAAUUGGCAAGGGACAUCCACUG ---------((((((.(....)(((((..((.........((((--(((.((((((((((((....)))))).))).))).).)))))).((((....))))))..))))))))))).. ( -34.10) >DroYak_CAF1 124716 113 - 1 ----UGUGGGUGGAUUUGGAUAGUUUUGGGUAUAUUAGGGUGGC--UGGAGCACUGGUGGUUUGGUGGCUACGCAGCUGCACGCAGCCGACAAUAGAAAUUGGCAAGGGACAUCCACUG ----.(((((((..((((..(((((((.....((((.(..((((--(((.((((((((((((....)))))).))).))).).)))))).)))))))))))).))))...))))))).. ( -39.00) >consensus GGUCUGUGGGUGGAUUUGUAUAGUUUUGGGUACAUUAGGGUGGCAGUGGAGCACUGGUGGUUUGGUGGCUACGCAGCUGCACGCAGCCGACAAUGGAAAUUGGCAAGGGACAUCCACUG .....(((((((..(((...(((((((.....((((.(..((((.(((..((((((((((((....)))))).))).))).))).)))).))))))))))))...)))..))))))).. (-34.26 = -35.10 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:52 2006