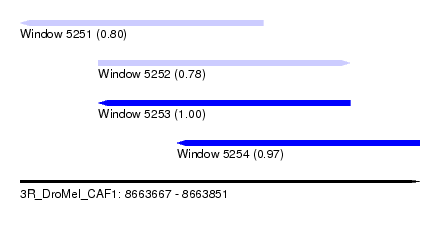
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,663,667 – 8,663,851 |
| Length | 184 |
| Max. P | 0.999921 |

| Location | 8,663,667 – 8,663,779 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.65 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8663667 112 - 27905053 AGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCU----CGCACAUGGUGCAAACGAAAA-GGCAAAGGCAU---UGGC ((((((((((........))))))))))..((((((((..........(((((....))))).(((....)))..)----)))))))(((((...(.....-)......))))---)... ( -29.50) >DroVir_CAF1 110809 110 - 1 AGGAAGCGAUAAAAUCAAAUUGUUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGG----CGCACAUGGUCUCAACA--UU-GGCAUCGGCAU---UGGC ((((((((((........))))))))))(((((((..((((((..........))))))..)))))))...(((((----(.......)))))).((--..-.((....))..---)).. ( -30.00) >DroWil_CAF1 112661 120 - 1 AGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGGAUGGAAGCACAUGGUGCAAGGGGAGAUGGUACAUACAUAUAUGGG ((((((((((........))))))))))..(((((((...........(....).(((((((.((((.......))))...((((...))))...)))))))....)))))))....... ( -32.90) >DroYak_CAF1 96492 111 - 1 AGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCU----CGCACAUGGUGCAAACGAAAA--GCAAAGGCAU---UGGC ((((((((((........))))))))))(((((((..((((((..........))))))..))))))).....(((----.((((...)))).........--((....))..---.))) ( -29.40) >DroMoj_CAF1 141265 110 - 1 AGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGG----CGCACAUGGUCUCAACA--GC-GGCAGAGACUU---GGGC ((((((((((........))))))))))(((((((..((((((..........))))))..))))))).......(----(.((...((((((....--..-....)))))))---).)) ( -32.80) >DroAna_CAF1 84675 112 - 1 AGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCC----CGCACAUGGUGCAAAAGGCAA-CUCAAAGGCAU---UGCC ((((((((((........))))))))))(((((((..((((((..........))))))..))))))).....(((----.((((...))))...((....-))....)))..---.... ( -33.70) >consensus AGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCG____CGCACAUGGUGCAAACG_AAA_GGCAAAGGCAU___UGGC ((((((((((........))))))))))(((((((..((((((..........))))))..))))))).............((((...))))............................ (-24.68 = -24.65 + -0.03)
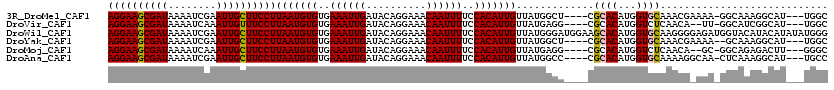
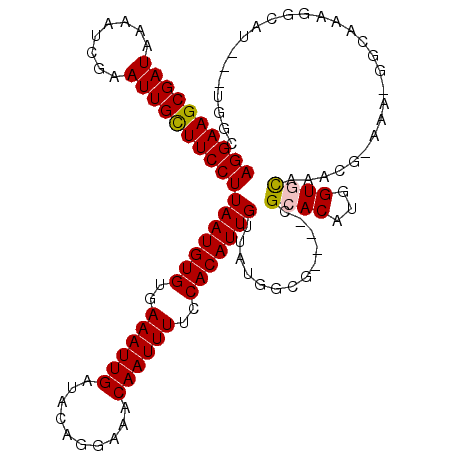
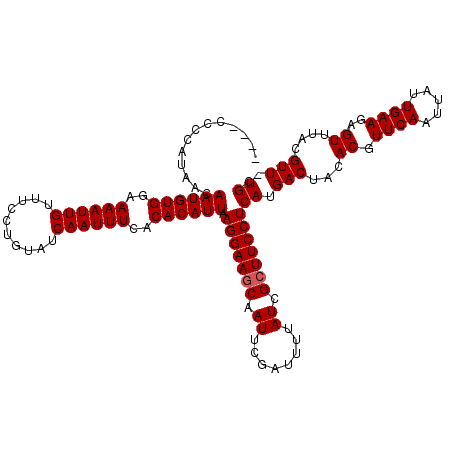
| Location | 8,663,703 – 8,663,819 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -20.13 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8663703 116 + 27905053 ----AGCCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAGCAAUUCGAUUUUAUCGCUUCCUCAUGACUAUACGUUCAAUUAUUGAAGAGUUUACGUUUUGC ----.........((((((..((((((..........))))))..)))))).(((((((.((........)).)))))))((.(((((.((.((((.....))))..)).)).))).)). ( -22.60) >DroVir_CAF1 110843 115 + 1 ----CCUCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAACAAUUUGAUUUUAUCGCUUCCUCAUGACUUCACGUUCAAUUAUUGAAGAGUUUACGUU-UGC ----.(((......(((.(((((((((((((((...................))))))))))))(((...)))...))).))).........((((.....)))))))........-... ( -20.31) >DroGri_CAF1 90099 115 + 1 ----CCUCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAGCAAUUUGAUUUUAUCGCUUCCUCAUGACUUCACGUUCAAUUAUUGAAGAGUUUACGUU-UGC ----..((((...((((((..((((((..........))))))..)))))).(((((((.((........)).))))))).))))(((((...........)))))..........-... ( -23.70) >DroWil_CAF1 112701 119 + 1 UCCAUCCCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAGCAAUUCGAUUUUAUCGCUUCCUCAUGACUACACGUUCAAUUAUUGAAGAGUUUACGUU-UGC .............((((((..((((((..........))))))..)))))).(((((((.((........)).)))))))...(((((.((.((((.....))))..)).)).)))-... ( -21.30) >DroMoj_CAF1 141299 115 + 1 ----CCUCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAGCAAUUUGAUUUUAUCGCUUCCUCAUGACUUCACGUUCAAUUAUUGAAGAGUUUACGUU-UGC ----..((((...((((((..((((((..........))))))..)))))).(((((((.((........)).))))))).))))(((((...........)))))..........-... ( -23.70) >DroAna_CAF1 84711 116 + 1 ----GGCCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAGCAAUUCGAUUUUAUCGCUUCCUCAUGACUACACGUUCAAUUAUUGAAGAGUUUACGUUUUGC ----.........((((((..((((((..........))))))..)))))).(((((((.((........)).)))))))((.(((((.((.((((.....))))..)).)).))).)). ( -22.40) >consensus ____CCCCAUAACAAUGUGGAAAAUUGUUUCCUGUAUCAAUUUCACACAUUAAGGAAGCAAUUCGAUUUUAUCGCUUCCUCAUGACUACACGUUCAAUUAUUGAAGAGUUUACGUU_UGC .............((((((..((((((..........))))))..)))))).(((((((.((........)).)))))))((.(((...((.((((.....))))..))....))).)). (-20.13 = -20.30 + 0.17)


| Location | 8,663,703 – 8,663,819 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -27.95 |
| Energy contribution | -27.56 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8663703 116 - 27905053 GCAAAACGUAAACUCUUCAAUAAUUGAACGUAUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCU---- .....((((.......(((.....)))))))..(((((((((((((((((........))))))))).(((((((..((((((..........))))))..)))))))))))))))---- ( -31.41) >DroVir_CAF1 110843 115 - 1 GCA-AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGUUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGG---- ...-..........(((((.........((((....))))((((((((((........))))))))))(((((((..((((((..........))))))..)))))))...)))))---- ( -27.50) >DroGri_CAF1 90099 115 - 1 GCA-AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGG---- ...-..........(((((.........((((....))))((((((((((........))))))))))(((((((..((((((..........))))))..)))))))...)))))---- ( -29.90) >DroWil_CAF1 112701 119 - 1 GCA-AACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGGAUGGA ...-..(((((.................((((....))))((((((((((........))))))))))(((((((..((((((..........))))))..))))))))))))....... ( -28.70) >DroMoj_CAF1 141299 115 - 1 GCA-AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGG---- ...-..........(((((.........((((....))))((((((((((........))))))))))(((((((..((((((..........))))))..)))))))...)))))---- ( -29.90) >DroAna_CAF1 84711 116 - 1 GCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCC---- ...........((.(((((.....)))).).)).((((((((((((((((........))))))))).(((((((..((((((..........))))))..)))))))))))))).---- ( -31.10) >consensus GCA_AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGG____ .....((((.......(((.....)))))))....(((((((((((((((........))))))))).(((((((..((((((..........))))))..)))))))))))))...... (-27.95 = -27.56 + -0.39)
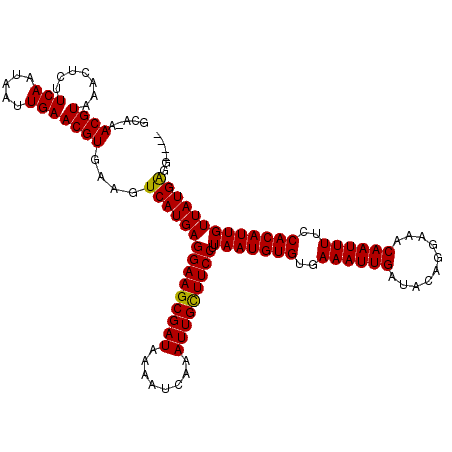
| Location | 8,663,739 – 8,663,851 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8663739 112 - 27905053 AGUGCUGGAAUUUCGGAAAG-------GGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUAUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAA ..((((((.....(((...(-------....).)))))))))..((((......((((.....))))..........((((((((((((........))))))))))))))))...... ( -29.70) >DroVir_CAF1 110879 112 - 1 AGUGGCGGACGGUGGCAAAA------GGAGGCGCAGACAGCA-AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGUUUCCUUAAUGUGUGAAA .((.((.....)).))....------....((((((((....-.((((.......(((.....)))))))...))).((((((((((((........)))))))))))).))))).... ( -25.81) >DroGri_CAF1 90135 106 - 1 ------------UGGGAAACGGAGUCGAAGGCGUAGACAGCA-AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAA ------------..(....)(((((((...((.......)).-..))...)))))(((...(((((.((((....)))).(((((((((........))))))))))))))...))).. ( -26.80) >DroWil_CAF1 112741 97 - 1 AUCA-----A--------CA-------UCAACG-CAACGGCA-AACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAA ....-----.--------..-------(((.((-((..(((.-.((((.......(((.....)))))))...))).((((((((((((........)))))))))))).))))))).. ( -25.81) >DroMoj_CAF1 141335 101 - 1 CGUGGU-----GCG------------GCAGGCGCAGACAGCA-AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAA .((..(-----(((------------....((.......)).-..)))).))..((((...(((((.((((....)))).(((((((((........))))))))))))))...)))). ( -26.80) >DroAna_CAF1 84747 112 - 1 AUCGCAGGAAUUUCAGGCCA-------ACAAGGCCGACGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAA .(((((.(.......((((.-------....))))...(((...((((.......(((.....)))))))...))).((((((((((((........)))))))))))).).))))).. ( -33.41) >consensus AGUG_____A__UGGGAAAA_______AAAACGCAGACAGCA_AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAA ................................((.....)).............((((...(((((.((((....)))).(((((((((........))))))))))))))...)))). (-19.10 = -19.18 + 0.08)

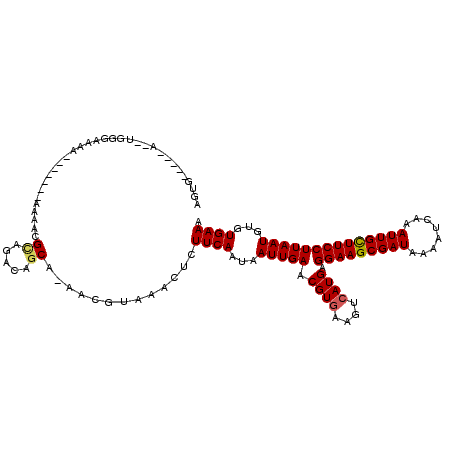
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:41 2006