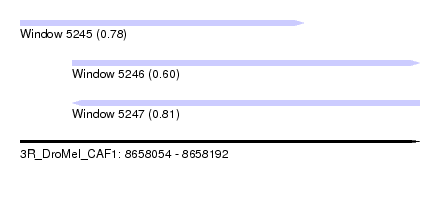
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,658,054 – 8,658,192 |
| Length | 138 |
| Max. P | 0.811786 |

| Location | 8,658,054 – 8,658,152 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -19.81 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8658054 98 + 27905053 CUUCGUCC-----GC-------CAUCGACCAUGCAAUGUUGUUCUUUCCGAUGUCGUUUGUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA ..(((...-----..-------...)))..(((((.(((.((...((((((((.(((..(((((.....))))).))).)))).))))...)))))..)))))....... ( -22.30) >DroVir_CAF1 106134 85 + 1 -------------------------CUGGCAUGCAAUGUUGUUCUUUCUGAUGUCGUUUAUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA -------------------------((((.(((((.(((.((..(((((((((.(((..(((((.....))))).))).)))))))))...)))))..))))).)))).. ( -25.20) >DroWil_CAF1 108080 110 + 1 UUCCACUCCGUCAGCCUUCCUUCAUCAGGUAUGCAAUGUUGUUCUUUCUGAUGUCGUUUGUUUUCGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA .............((...(((.....))).(((((.(((.((..(((((((((.(((....((((....))))..))).)))))))))...)))))..)))))....)). ( -26.60) >DroMoj_CAF1 136322 85 + 1 -------------------------GUGCCUGCCAAUGUUGUUCUUUCUGAUGUCGUUUAUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA -------------------------(..((((.......(((..(((((((((.(((..(((((.....))))).))).)))))))))...)))))))..)......... ( -24.61) >DroAna_CAF1 79505 95 + 1 UUCCAC--------C-------CACAGACCAUGCAAUGUUGUUCUUUCCGAUGUCGUUUGUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA ......--------.-------........(((((.(((.((...((((((((.(((..(((((.....))))).))).)))).))))...)))))..)))))....... ( -21.40) >DroPer_CAF1 92271 103 + 1 AUUCGCCCACUCACC-------CUUCGAGCAUGCAAUGUUGUUCUUUCUGAUGUCGUUUGUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA ....((...(((...-------....))).(((((.(((.((..(((((((((.(((..(((((.....))))).))).)))))))))...)))))..)))))....)). ( -25.90) >consensus _U_C_C________C_______CAUCGACCAUGCAAUGUUGUUCUUUCUGAUGUCGUUUGUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCA ..............................(((((.(((.((..(((((((((.(((..(((((.....))))).))).)))))))))...)))))..)))))....... (-19.81 = -20.42 + 0.61)



| Location | 8,658,072 – 8,658,192 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -24.87 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8658072 120 + 27905053 AUGCAAUGUUGUUCUUUCCGAUGUCGUUUGUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCUGGCAAUUUAAUGCUGUUGUUC ..(((((..(((...((((((((.(((..(((((.....))))).))).)))).))))...)))...((((......))))............))))).((((......))))....... ( -27.40) >DroVir_CAF1 106139 120 + 1 AUGCAAUGUUGUUCUUUCUGAUGUCGUUUAUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCUGGCAAUUUAAUACAGUUGUUA ..(((((..(((..(((((((((.(((..(((((.....))))).))).)))))))))...)))...((((......))))............)))))((((((((......)))))))) ( -29.20) >DroGri_CAF1 85705 120 + 1 AUGCAAUGUUGUUCUUUCUGAUGUCGUUUAUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCUGGCAAUUUAAUACAGUUGUUA ..(((((..(((..(((((((((.(((..(((((.....))))).))).)))))))))...)))...((((......))))............)))))((((((((......)))))))) ( -29.20) >DroWil_CAF1 108110 120 + 1 AUGCAAUGUUGUUCUUUCUGAUGUCGUUUGUUUUCGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCUAGCAAUUUAAUGCUGCUCUUG ..(((((..(((..(((((((((.(((....((((....))))..))).)))))))))...)))...((((......))))............)))))(((((......)))))...... ( -31.10) >DroMoj_CAF1 136327 120 + 1 UGCCAAUGUUGUUCUUUCUGAUGUCGUUUAUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCUGGCAAUUUAAUACAGUUGUUA (((((.(((.((..(((((((((.(((..(((((.....))))).))).)))))))))...))))).((((......)))).................)))))................. ( -29.20) >DroAna_CAF1 79520 120 + 1 AUGCAAUGUUGUUCUUUCCGAUGUCGUUUGUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCCGGCAAUUUAAUGCUGUUGUUC ..(((((..(((...((((((((.(((..(((((.....))))).))).)))).))))...)))...((((......))))............)))))(((((......)))))...... ( -28.90) >consensus AUGCAAUGUUGUUCUUUCUGAUGUCGUUUAUUUUUGUUCGAAAUUGCGCCAUCAGGAAUAUGCACAGGUGCAUUUUAGCACAAUAAAAACGUAAUUGCUGGCAAUUUAAUACAGUUGUUA ..(((((..(((..(((((((((.(((..(((((.....))))).))).)))))))))...)))...((((......))))............))))).((((((........)))))). (-24.87 = -25.32 + 0.45)


| Location | 8,658,072 – 8,658,192 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -20.89 |
| Energy contribution | -20.58 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8658072 120 - 27905053 GAACAACAGCAUUAAAUUGCCAGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACAAAAACAAACGACAUCGGAAAGAACAACAUUGCAU ........(((......)))..(((((...(((((((((((((((..(((((....)))))........)))))))))(((((................)))))))))))...))))).. ( -24.09) >DroVir_CAF1 106139 120 - 1 UAACAACUGUAUUAAAUUGCCAGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACAAAAAUAAACGACAUCAGAAAGAACAACAUUGCAU ......................(((((...((((((...((((......))))............((((((.((..((((.......))))...)).)))))).))))))...))))).. ( -21.00) >DroGri_CAF1 85705 120 - 1 UAACAACUGUAUUAAAUUGCCAGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACAAAAAUAAACGACAUCAGAAAGAACAACAUUGCAU ......................(((((...((((((...((((......))))............((((((.((..((((.......))))...)).)))))).))))))...))))).. ( -21.00) >DroWil_CAF1 108110 120 - 1 CAAGAGCAGCAUUAAAUUGCUAGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACGAAAACAAACGACAUCAGAAAGAACAACAUUGCAU .......((((......)))).(((((...((((((...((((......))))............((((((.((...((((....)))).....)).)))))).))))))...))))).. ( -27.20) >DroMoj_CAF1 136327 120 - 1 UAACAACUGUAUUAAAUUGCCAGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACAAAAAUAAACGACAUCAGAAAGAACAACAUUGGCA .................((((((.......((((..(((((((((..(((((....)))))........)))))))))...))))..............((.....))......)))))) ( -21.40) >DroAna_CAF1 79520 120 - 1 GAACAACAGCAUUAAAUUGCCGGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACAAAAACAAACGACAUCGGAAAGAACAACAUUGCAU ......(.(((......))).)(((((...(((((((((((((((..(((((....)))))........)))))))))(((((................)))))))))))...))))).. ( -24.29) >consensus UAACAACAGCAUUAAAUUGCCAGCAAUUACGUUUUUAUUGUGCUAAAAUGCACCUGUGCAUAUUCCUGAUGGCGCAAUUUCGAACAAAAACAAACGACAUCAGAAAGAACAACAUUGCAU ........(((......)))..(((((...((((((...((((......))))............((((((.((....................)).)))))).))))))...))))).. (-20.89 = -20.58 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:34 2006