
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 8,650,426 – 8,650,546 |
| Length | 120 |
| Max. P | 0.683421 |

| Location | 8,650,426 – 8,650,520 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -10.85 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8650426 94 + 27905053 CAAUGUAUUCUUGGGU-UAUAAAAAUCAGGGUGGAAACCAAAAAAAAAAAAAAAAAAACCGAAAAUCGA----------GCAAUUGCUUAAAGUUGCUGGUUCUA ........((((((..-........))))))(((((.((....................((.....))(----------((((((......)))))))))))))) ( -14.80) >DroSec_CAF1 77303 91 + 1 CAAUGUAUUUUUAGGC-UAUAAAAAUCAGGGUGGAAUCCCAAAAAAAAA---AUGAAACCGAAAAUGGA----------GUAACUGCUUAAAGUUGCUGGCUCUA ...........(((((-(..........(((......))).........---......((......))(----------((((((......)))))))))).))) ( -16.10) >DroSim_CAF1 93575 89 + 1 CAAUGUAUUUUUAGGC-UAUAAAAAUCAGGGUGGAAACCCAAAAAAAAA-----GAAACCGAAAAUGGA----------GCACCUGCUUAAAGUUGCUGGCUCUA ...((.(((((((...-..)))))))))((((....)))).........-----...........((((----------((....((........))..)))))) ( -20.20) >DroYak_CAF1 82749 102 + 1 CAAUGUAUUUUUGUGUCCUUAAAAAUGUGGCUAGAAAGCCAAAAAAAAA---CAAGAAACGAGAAUGGAGCCACCGAAGGCAACUGCUUAAAGUUGCCAGCUCUA ......((((((((.((..........(((((....)))))........---...)).))))))))(((((.......(((((((......))))))).))))). ( -27.40) >consensus CAAUGUAUUUUUAGGC_UAUAAAAAUCAGGGUGGAAACCCAAAAAAAAA___AAAAAACCGAAAAUGGA__________GCAACUGCUUAAAGUUGCUGGCUCUA ......(((((((......)))))))..((((....)))).......................................((((((......))))))........ (-10.85 = -11.60 + 0.75)



| Location | 8,650,453 – 8,650,546 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -14.76 |
| Energy contribution | -16.57 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 8650453 93 + 27905053 GGGUGGAAACCAAAAAAAAAAAAAAAAAAACCGAAAAUCGA----------GCAAUUGCUUAAAGUUGCUGGUUCUAAUUAUUGUCCUCGUUCCCGGUUGUGA .(((....))).................(((((..((.(((----------((((((......)))))).((..(........).)))))))..))))).... ( -17.80) >DroSec_CAF1 77330 90 + 1 GGGUGGAAUCCCAAAAAAAAA---AUGAAACCGAAAAUGGA----------GUAACUGCUUAAAGUUGCUGGCUCUAAUUAUUGUCCUCGUUCUCGGUUGUGA (((......))).........---....((((((.((((((----------((((((......)))))))(((..........))).))))).)))))).... ( -23.10) >DroSim_CAF1 93602 88 + 1 GGGUGGAAACCCAAAAAAAAA-----GAAACCGAAAAUGGA----------GCACCUGCUUAAAGUUGCUGGCUCUAAUUAUUGUCCUCGUUCUCGGUUGUGA ((((....)))).........-----..((((((.((((((----------(((.((......)).))))(((..........))).))))).)))))).... ( -24.30) >DroYak_CAF1 82777 100 + 1 GGCUAGAAAGCCAAAAAAAAA---CAAGAAACGAGAAUGGAGCCACCGAAGGCAACUGCUUAAAGUUGCCAGCUCUAAUUAUUGUCCUCGUUCUCAGUUGUGA ((((....)))).........---((...((((((((((((((.......(((((((......))))))).)))).............))))))).))).)). ( -26.91) >consensus GGGUGGAAACCCAAAAAAAAA___AAAAAACCGAAAAUGGA__________GCAACUGCUUAAAGUUGCUGGCUCUAAUUAUUGUCCUCGUUCUCGGUUGUGA ((((....))))................((((((.(((((...........((((((......)))))).(((..........))).))))).)))))).... (-14.76 = -16.57 + 1.81)

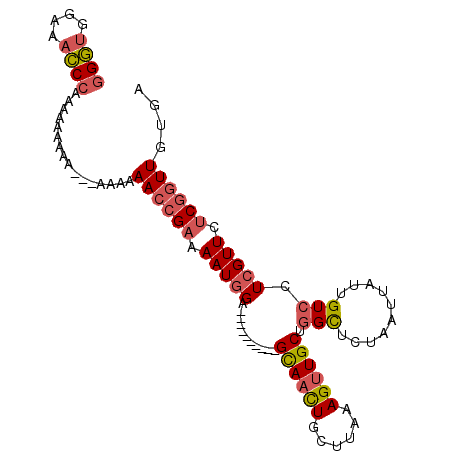
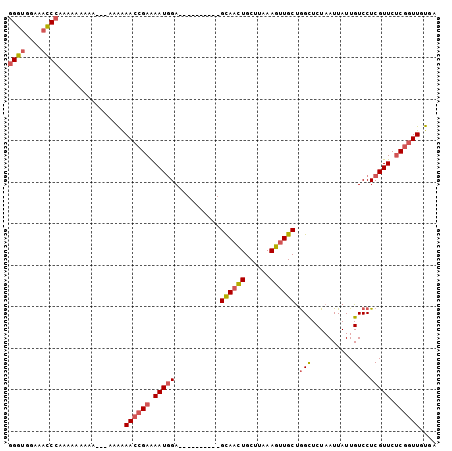
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:26 2006